The LBP Gene and Its Association with Resistance to Aeromonas hydrophila in Tilapia
Abstract
:1. Introduction
2. Results
2.1. Sequence of cDNA and Genomic DNA of the Lipopolysaccharide Binding Protein (LBP) Gene
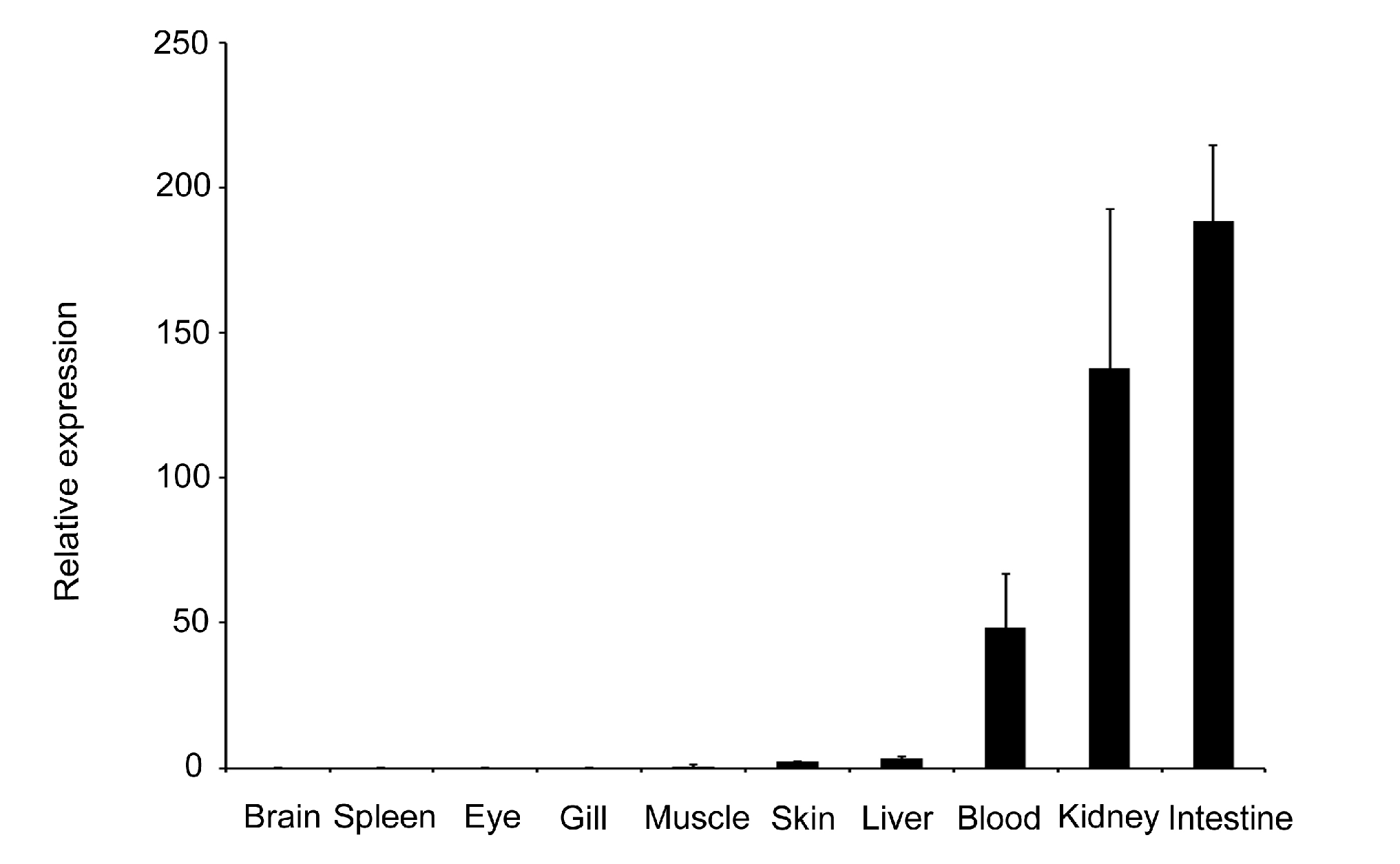
2.2. Expression Profiles of the LBP Gene in Normal Individuals
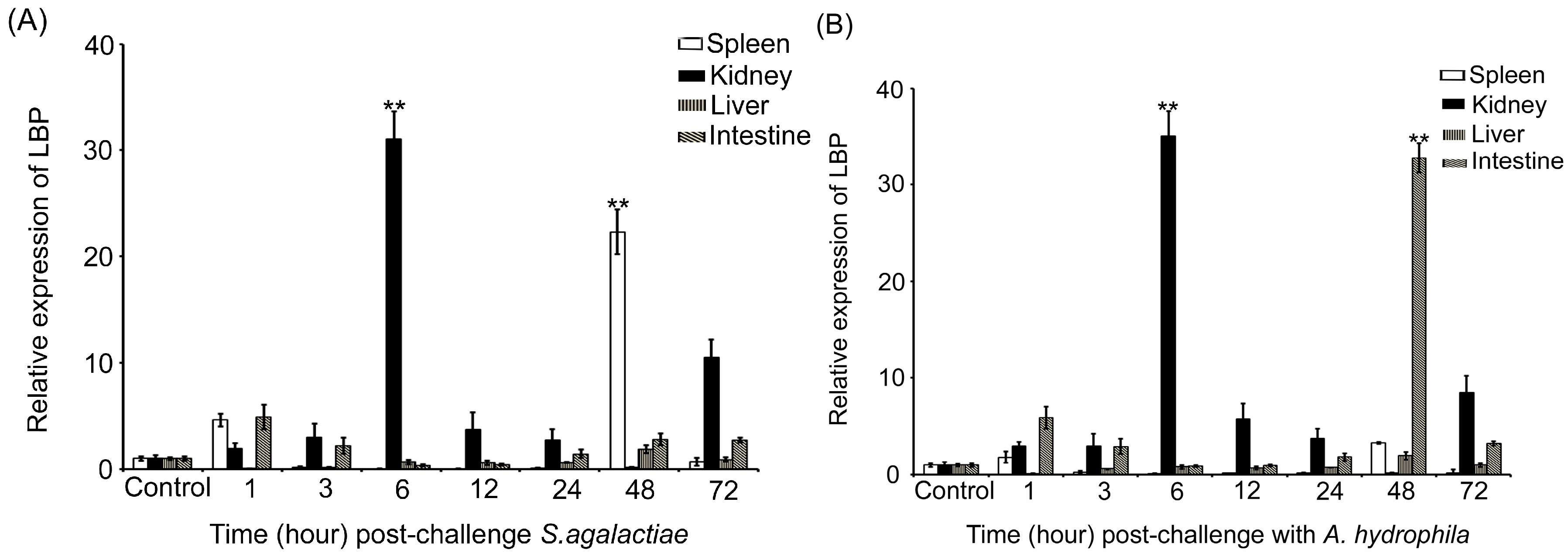
2.3. Expression Profiles of the LBP Gene in Individuals Infected with Bacteria
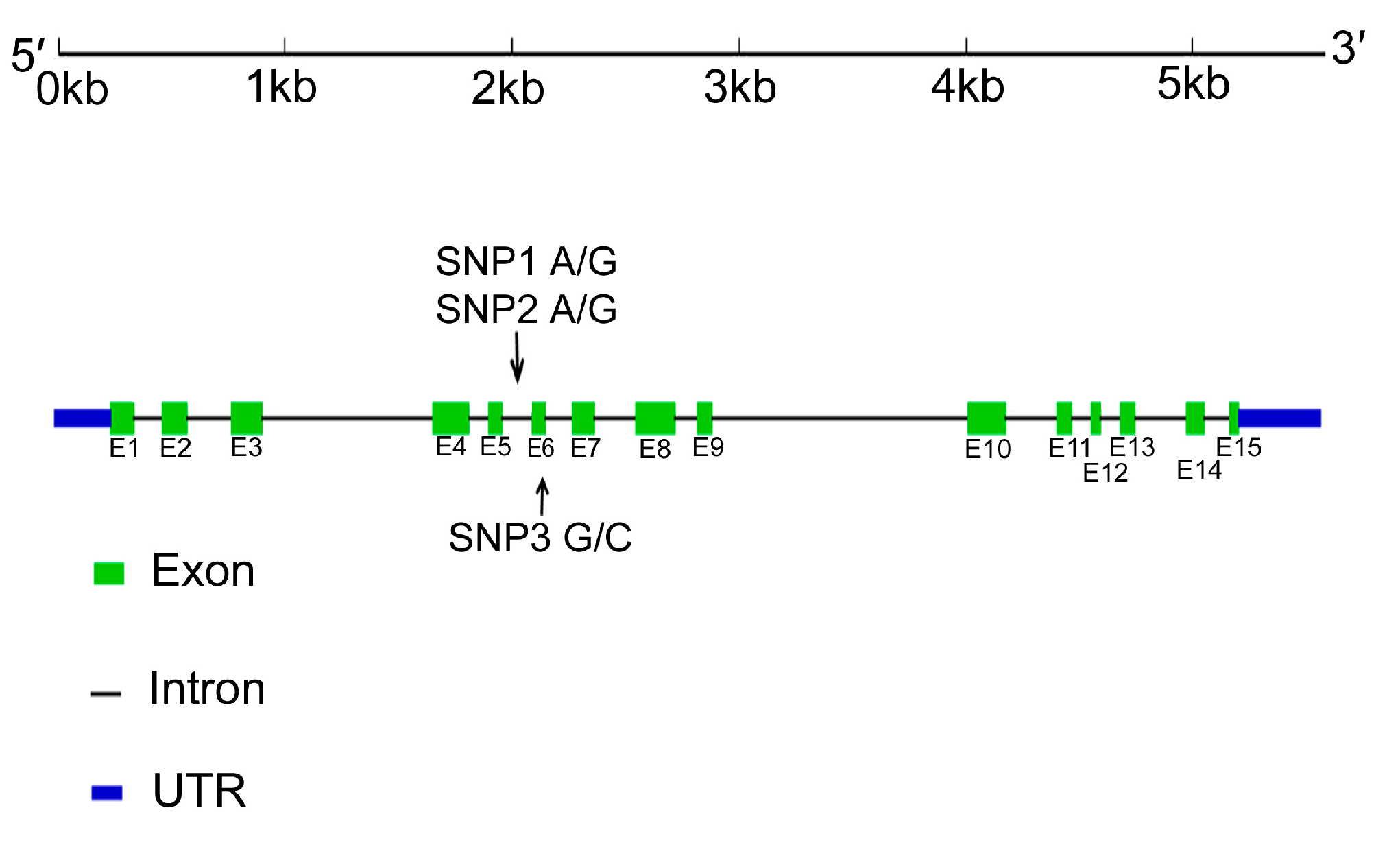
2.4. Identification of Single Nucleotide Polymorphisms (SNPs) in the LBP Gene
2.5. Identification of Associations between SNPs and Disease Resistance/Susceptibility to Bacteria in Tilapia
| SNP | Number (%) | Allele Number (%) | |||||
|---|---|---|---|---|---|---|---|
| Genotypes | Susceptible | Resistant | X2 | Allele | Susceptible | Resistant | X2 |
| S. agalactiae | |||||||
| SNP1 | |||||||
| AA ( n = 332) | 200 (66) | 132 (73) | 2.03 | A | 202 (66) | 114 (70) | 1.12 |
| AG ( n = 150) | 102 (34) | 48 (27) | ( p > 0.05) | G | 102 (34) | 48 (30) | ( p > 0.05) |
| GG ( n = 0) | 0 (0) | 0 (0) | |||||
| SNP2 | |||||||
| AA ( n = 8) | 6 (2) | 2 (1) | 1.14 | A | 86 (20) | 38 (16) | 2.26 |
| AG ( n = 58) | 40 (13) | 18 (10) | ( p > 0.05) | G | 340 (80) | 196 (84) | ( p > 0.05) |
| GG ( n = 420) | 260 (85) | 160 (89) | |||||
| CC ( n = 18) | 14 (5) | 4 (2) | 1.19 | C | 134 (28) | 76 (26) | 0.29 |
| CG ( n = 96) | 60 (20) | 36 (20) | ( p > 0.05) | G | 348 (72) | 212 (74) | ( p > 0.05) |
| GG ( n = 368) | 228 (75) | 140 (78) | |||||
| A. hydrophila | |||||||
| SNP1 | |||||||
| AA ( n = 354) | 188 (71) | 166 (88) | 14.61 | A | 340 (69) | 206 (82) | 20.70 |
| AG ( n = 96) | 76 (59) | 20 (11) | ( p < 0.005) | G | 152 (31) | 44 (18) | ( p < 0.05) |
| GG ( n = 2) | 0 (0) | 2 (1) | |||||
| SNP2 | |||||||
| AA ( n = 8) | 8 (3) | 0 (0) | 2.97 | A | 52 (16) | 32 (13) | 0.96 |
| AG ( n = 38) | 22 (8) | 16 (8) | ( p > 0.05) | G | 278 (84) | 206 (87) | ( p > 0.05) |
| GG ( n = 408) | 234 (89) | 174 (92) | |||||
| SNP3 | |||||||
| CC ( n = 28) | 8 (3) | 20 (10) | 18.34 | C | 100 (25) | 72 (27) | 0.50 |
| CG ( n = 72) | 46 (18) | 26 (14) | ( p < 0.005) | G | 300 (75) | 196 (73) | ( p > 0.05) |
| GG ( n = 352) | 208 (79) | 144 (76) | |||||
2.6. Associations between SNPs in the LBP Gene and Growth Traits
2.7. In-Silico Mapping of the LPB Gene to Linkage Group 20
3. Discussion
4. Experimental Section
4.1. Fish and Ethics Statement
4.2. Identification of cDNA and Genomic DNA Sequences of the LBP Gene
| Name | Primer Sequence (5'–3') | T (°C) | Product Length (bp) | Application |
|---|---|---|---|---|
| LBP-RT-F1 | GGCGCAGCTGGGGAAAGAA | 60 | 269 | qRT-PCR |
| LBP-RT-R1 | TGGGGACATCAGTGAGAGGAAGG | |||
| β-actin F1 | TGACCCAGATCATGTTCGAGAC | 60 | 253 | qRT-PCR |
| β-actin R1 | GTGGTGGTGAAGGAGTAGCC | |||
| LBP-G-F1 | ACTTCTCAGTGACACAGGAAATTA | 57 | 1827 | Genomic DNA |
| LBP-G-R1 | CTGGCTCCACCATGAAATTCTAT | |||
| LBP-G-F3 | GAAAAGCAAAACCAACCAGCTTG | 57 | 2004 | Genomic DNA |
| LBP-G-R3 | TATGACAGAAGTTGTTTTTAATCCT | |||
| LBP-G-F2 | GGCGCAGCTGGGGAAAGAAGCTGA | 57 | 1902 | SNP detection |
| LBP-G-R2 | TAGTTATAGGCATAGTATATGTTTG |
4.3. RNA Extraction and Quantitative Real-Time RT-PCR
4.4. Bacterial Challenge and Sampling Tissues
4.5. Identification and Genotyping of SNPs in the LBP Gene
4.6. Association Analysis of SNPs with Resistance to the Infection of Two Bacterial Pathogens
4.7. Association Analysis of SNPs in the LBP Gene with Growth Traits
4.8. Mapping the LBP Gene to the Genome of Tilapia
Abbreviations
| LBP | lipopolysaccharide-binding protein |
| S. Agalactiae | Streptcoccus agalactiae |
| A. Hydrophila | Aeromonas hydrophila |
| SNP | Single nucleotide polymorphism |
| qRT-PCR | Quantitative real-time PCR |
Supplementary Material
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Austin, B.; Austin, D.; Munn, C. Bacterial fish pathogens: Disease in farmed and wild fish. Rev. Fish. Biol. Fish. 1995, 5, 123–124. [Google Scholar]
- Duremdez, R.; Al-Marzouk, A.; Qasem, J.; Al-Harbi, A.; Gharabally, H. Isolation of Streptococcus agalactiae from cultured silver pomfret, Pampus argenteus (euphrasen), in kuwait. J. Fish Dis. 2004, 27, 307–310. [Google Scholar]
- Mian, G.; Godoy, D.; Leal, C.; Yuhara, T.; Costa, G.; Figueiredo, H. Aspects of the natural history and virulence of S. agalactiae infection in nile tilapia. Vet. Microbiol. 2009, 136, 180–183. [Google Scholar]
- Evans, J.; Klesius, P.; Gilbert, P.; Shoemaker, C.; Al Sarawi, M.; Landsberg, J.; Duremdez, R.; Al Marzouk, A.; Al Zenki, S. Characterization of β-haemolytic group b Streptococcus agalactiae in cultured seabream, Sparus auratus L., and wild mullet, liza klunzingeri (day), in kuwait. J. Fish Dis. 202, 25, 505–513. [Google Scholar]
- Zamri-Saad, M.; Amal, M.; Siti-Zahrah, A. Pathological changes in red tilapias (Oreochromis spp.) naturally infected by Streptococcus agalactiae. J. Comp. Pathol. 2010, 143, 227–229. [Google Scholar]
- Rodriguez, I.; Novoa, B.; Figueras, A. Immune response of zebrafish (Danio rerio) against a newly isolated bacterial pathogen Aeromonas hydrophila. Fish Shellfish Immunol. 2008, 25, 239–249. [Google Scholar]
- Li, A.; Yang, W.; Hu, J.; Wang, W.; Cai, T.; Wang, J. Optimization by orthogonal array design and humoral immunity of the bivalent vaccine against Aeromonas hydrophila and Vibrio fluvialis infection in crucian carp (Carassius auratus L.). Aquat. Res. 2006, 37, 813–820. [Google Scholar]
- Ardó, L.; Yin, G.; Xu, P.; Váradi, L.; Szigeti, G.; Jeney, Z.; Jeney, G. Chinese herbs (Astragalus membranaceus and Lonicera japonica) and boron enhance the non-specific immune response of nile tilapia (Oreochromis niloticus) and resistance against Aeromonas hydrophila. Aquaculture 2008, 275, 26–33. [Google Scholar]
- Yue, G.H. Recent advances of genome mapping and marker-assisted selection in aquaculture. Fish Fish. 2014, 15, 376–396. [Google Scholar]
- Houston, R.D.; Haley, C.S.; Hamilton, A.; Guy, D.R.; Tinch, A.E.; Taggart, J.B.; McAndrew, B.J.; Bishop, S.C. Major quantitative trait loci affect resistance to infectious pancreatic necrosis in atlantic salmon (Salmo salar). Genetics 2008, 178, 1109–1115. [Google Scholar]
- Fuji, K.; Hasegawa, O.; Honda, K.; Kumasaka, K.; Sakamoto, T.; Okamoto, N. Marker-assisted breeding of a lymphocystis disease-resistant Japanese flounder (Paralichthys olivaceus). Aquaculture 2007, 272, 291–295. [Google Scholar]
- Vignal, A.; Milan, D.; SanCristobal, M.; Eggen, A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 2002, 34, 275–306. [Google Scholar]
- Lazarus, R.; Vercelli, D.; Palmer, L.J.; Klimecki, W.J.; Silverman, E.K.; Richter, B.; Riva, A.; Ramoni, M.; Martinez, F.D.; Weiss, S.T. Single nucleotide polymorphisms in innate immunity genes: Abundant variation and potential role in complex human disease. Immunol. Rev. 2002, 190, 9–25. [Google Scholar]
- Kongchum, P.; Sandel, E.; Lutzky, S.; Hallerman, E.M.; Hulata, G.; David, L.; Palti, Y. Association between IL-10a single nucleotide polymorphisms and resistance to cyprinid herpesvirus-3 infection in common carp (Cyprinus carpio). Aquaculture 2011, 315, 417–421. [Google Scholar]
- Siva, V.S.; Yang, C.; Yang, J.; Wang, L.; Wang, L.; Zhou, Z.; Qiu, L.; Song, L. Association of lgbp gene polymorphism with disease susceptibility/resistance of zhikong scallop (Chlamys farreri) to Listonella anguillarum. Fish Shellfish Immunol. 2012, 32, 1117–1123. [Google Scholar]
- Fu, G.H.; Bai, Z.Y.; Xia, J.H.; Liu, X.J.; Liu, F.; Wan, Z.Y.; Yue, G.H. Characterization of the LECT2 gene and its associations with resistance to the big belly disease in asian seabass. Fish Shellfish Immunol. 2014, 37, 131–138. [Google Scholar]
- Fu, G.H.; Bai, Z.Y.; Xia, J.H.; Liu, F.; Liu, P.; Yue, G.H. Analysis of two lysozyme genes and antimicrobial functions of their recombinant proteins in Asian seabass. PLoS One 2013, 8, e79743. [Google Scholar]
- Schumann, R.R.; Flaggs, G.; Gray, P.; Wright, S.; Mathison, J.; Tobias, P.; Ulevitch, R. Structure and function of lipopolysaccharide binding protein. Science 1990, 249, 1429–1431. [Google Scholar]
- Weiss, J. Bactericidal/permeability-increasing protein (bpi) and lipopolysaccharide-binding protein (lbp): Structure, function and regulation in host defence against gram-negative bacteria. Biochem. Soc. Trans. 2003, 31, 785–790. [Google Scholar]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Ann. Rev. Biochem. 2002, 71, 635. [Google Scholar]
- Watson, J.; Riblet, R. Genetic control of responses to bacterial lipopolysaccharides in mice. I. Evidence for a single gene that influences mitogenic and immunogenic respones to lipopolysaccharides. J. Exp. Med. 1974, 140, 1147–1161. [Google Scholar]
- Baba, T.; Imamura, J.; Izawa, K.; Ikeda, K. Immune protection in carp, Cyprinus carpio L., after immunization with Aeromonas hydrophila crude lipopolysaccharide. J. Fish Dis. 1988, 11, 237–244. [Google Scholar]
- Guttvik, A.; Paulsen, B.; Dalmo, R.A.; Espelid, S.; Lund, V.; Bøgwald, J. Oral administration of lipopolysaccharide to atlantic salmon (Salmo salar L.) fry. Uptake, distribution, influence on growth and immune stimulation. Aquaculture 2002, 214, 35–53. [Google Scholar]
- El-Sayed, A.-F.M. Tilapia Culture; CABI: Oxford, UK, 2006. [Google Scholar]
- Liu, F.; Sun, F.; Li, J.; Xia, J.H.; Lin, G.; Tu, R.J.; Yue, G.H. A microsatellite-based linkage map of salt tolerant tilapia (Oreochromis mossambicus × Oreochromis spp.) and mapping of sex-determining loci. BMC Genomics 2013, 14, 58. [Google Scholar]
- Xia, J.H.; Lin, G.; He, X.; Liu, P.; Liu, F.; Sun, F.; Tu, R.; Yue, G.H. Whole genome scanning and association mapping identified a significant association between growth and a SNP in the IFABP—A gene of the asian seabass. BMC Genomics 2013, 14, 295. [Google Scholar]
- Su, G.L.; Freeswick, P.D.; Geller, D.A.; Wang, Q.; Shapiro, R.A.; Wan, Y.-H.; Billiar, T.R.; Tweardy, D.J.; Simmons, R.L.; Wang, S.C. Molecular cloning, characterization, and tissue distribution of rat lipopolysaccharide binding protein. Evidence for extrahepatic expression. J. Immunol. 1994, 153, 743–752. [Google Scholar]
- Kim, J.W.; Gerwick, L.; Park, C.I. Molecular identification and expression analysis of two distinct bpi/lbps (bactericidal permeability-increasing protein/lps-binding protein) from rock bream, Oplegnathus fasciatus. Fish Shellfish Immunol. 2012, 33, 75–84. [Google Scholar]
- Lu, X.J.; Chu, C.Q.; Chen, Q.; Chen, J. A novel lipopolysaccharide-binding protein (LBP) gene from sweetfish Plecoglossus altivelis: Molecular characterization and its role in the immune response of monocytes/macrophages. Fish Shellfish Immunol. 2014, 38, 111–118. [Google Scholar]
- Inagawa, H.; Honda, T.; Kohchi, C.; Nishizawa, T.; Yoshiura, Y.; Nakanishi, T.; Yokomizo, Y.; Soma, G.I. Cloning and characterization of the homolog of mammalian lipopolysaccharide-binding protein and bactericidal permeability-increasing protein in rainbow trout Oncorhynchus mykiss. J. Immunol. 2002, 168, 5638–5644. [Google Scholar]
- Larrick, J.W.; Hirata, M.; Balint, R.F.; Lee, J.; Zhong, J.; Wright, S.C. Human cap18: A novel antimicrobial lipopolysaccharide-binding protein. Infect. Immun. 1995, 63, 1291–1297. [Google Scholar]
- Hubacek, J.A.; Büchler, C.; Aslanidis, C.; Schmitz, G. The genomic organization of the genes for human lipopolysaccharide binding protein (LBP) and bactericidal permeability increasing protein (BPI) is highly conserved. Biochem. Biophys. Res. Commun. 1997, 236, 427–430. [Google Scholar]
- Kirschning, C.J.; Au-Young, J.; Lamping, N.; Reuter, D.; Pfeil, D.; Seilhamer, J.J.; Schumann, R.R. Similar organization of the lipopolysaccharide-binding protein (LBP) and phospholipid transfer protein (PLTP) genes suggests a common gene family of lipid-binding proteins. Genomics 1997, 46, 416–425. [Google Scholar]
- Kono, T.; Sakai, M. Molecular cloning of a novel bactericidal permeability-increasing protein/lipopolysaccharide-binding protein (BPI/LBP) from common carp Cyprinus carpio L. and its expression. Mol. Immunol. 2003, 40, 269–278. [Google Scholar]
- Press, C.M.; Evensen, Ø. The morphology of the immune system in teleost fishes. Fish Shellfish Immunol. 1999, 9, 309–318. [Google Scholar]
- Insoft, R.M.; Sanderson, I.R.; Walker, W.A. Development of immune function in the intestine and its role in neonatal diseases. Pediatr. Clin. N. Am. 1996, 43, 551–571. [Google Scholar]
- Uematsu, S.; Fujimoto, K. The innate immune system in the intestine. Microbiol. Immun. 2010, 54, 645–657. [Google Scholar]
- Toranzo, A.E.; Magariños, B.; Romalde, J.L. A review of the main bacterial fish diseases in mariculture systems. Aquaculture 2005, 246, 37–61. [Google Scholar]
- Woo, P.T.; Bruno, D.W. Fish Diseases and Disorders: Viral, Bacterial and Fungal Infections; CABI: Oxford, UK, 2011; Volume 3. [Google Scholar]
- Klein, R.D.; Su, G.L.; Schmidt, C.; Aminlari, A.; Steinstraesser, L.; Alarcon, W.H.; Zhang, H.Y.; Wang, S.C. Lipopolysaccharide-binding potein acelerates and augments Escherichia coli phagocytosis by alveolar macrophages. J. Surg. Res. 2000, 94, 159–166. [Google Scholar]
- Schröder, N.W.; Heine, H.; Alexander, C.; Manukyan, M.; Eckert, J.; Hamann, L.; Göbel, U.B.; Schumann, R.R. Lipopolysaccharide binding protein binds to triacylated and diacylated lipopeptides and mediates innate immune responses. J. Immunol. 2004, 173, 2683–2691. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar]
- Yue, G.H.; Orban, L. A simple and affordable method for high-throughput DNA extraction from animal tissues for polymerase chain reaction. Electrophoresis 2005, 26, 3081–3083. [Google Scholar]
- Barrett, J.C.; Orban, L. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb. Protoc. 2009. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, G.H.; Liu, F.; Xia, J.H.; Yue, G.H. The LBP Gene and Its Association with Resistance to Aeromonas hydrophila in Tilapia. Int. J. Mol. Sci. 2014, 15, 22028-22041. https://doi.org/10.3390/ijms151222028
Fu GH, Liu F, Xia JH, Yue GH. The LBP Gene and Its Association with Resistance to Aeromonas hydrophila in Tilapia. International Journal of Molecular Sciences. 2014; 15(12):22028-22041. https://doi.org/10.3390/ijms151222028
Chicago/Turabian StyleFu, Gui Hong, Feng Liu, Jun Hong Xia, and Gen Hua Yue. 2014. "The LBP Gene and Its Association with Resistance to Aeromonas hydrophila in Tilapia" International Journal of Molecular Sciences 15, no. 12: 22028-22041. https://doi.org/10.3390/ijms151222028





