De Novo Assembly and Characterization of Pericarp Transcriptome and Identification of Candidate Genes Mediating Fruit Cracking in Litchi chinensis Sonn.
Abstract
:1. Introduction
2. Results
2.1. cDNA Sequence Generation and de Novo Assembly
| Category | Sample 1 | Sample 2 |
|---|---|---|
| Number of reads | 25,867,380 | 29,444,512 |
| Average read length (bp) | 100 | 100 |
| Total length of reads (bp) | 2,586,738,000 | 2,944,451,200 |
| Number of contigs | 42,790 | 42,447 |
| Average length of contigs (bp) | 966 | 967 |
| Number of unigenes | 38,917 | 38,695 |
| Average length of unigenes (bp) | 949 | 950 |
| Total length of unigenes | 36,945,798 | 36,761,221 |
2.2. Functional Annotation of the Transcriptome in Litchi Pericarp
| Public Protein Database | Number of Unigene Hits | Percentage (%) * |
|---|---|---|
| NCBI NR | 32,479 | 69.64 |
| Swiss-Prot | 25,010 | 53.62 |
| COG | 12,981 | 27.83 |
| KEGG | 9504 | 20.38 |

2.3. GO Annotation
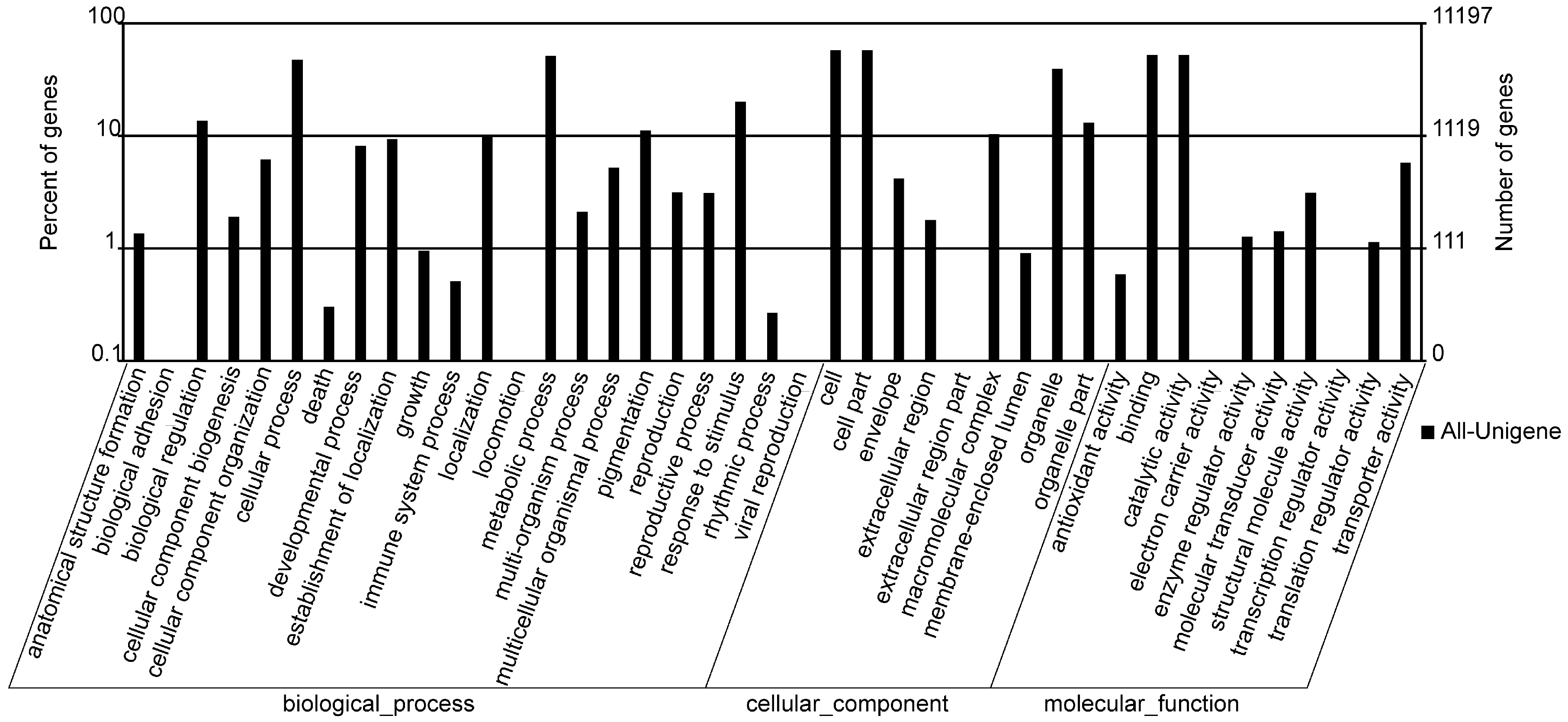
2.4. COG Annotation
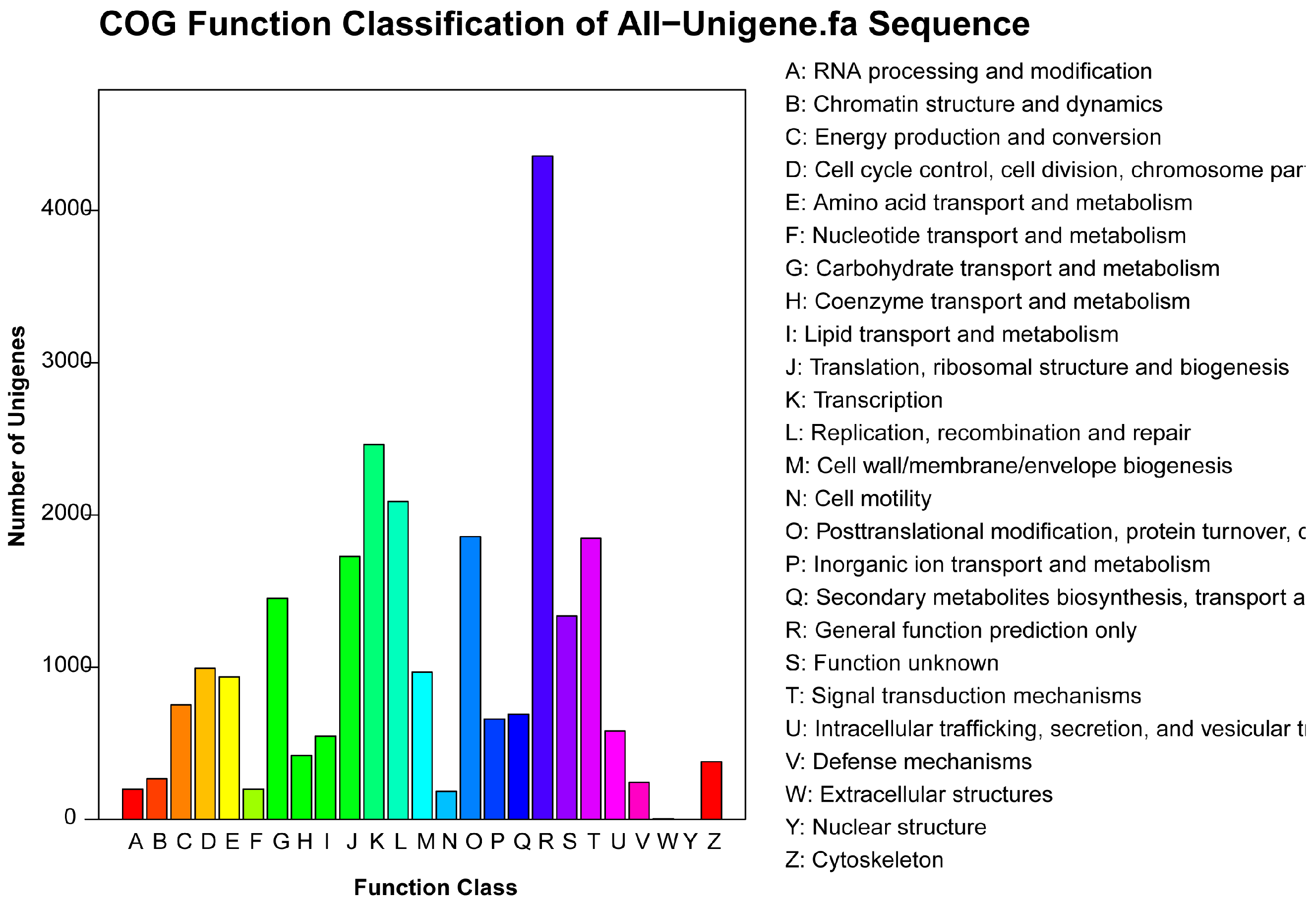
2.5. KEGG Pathway Mapping
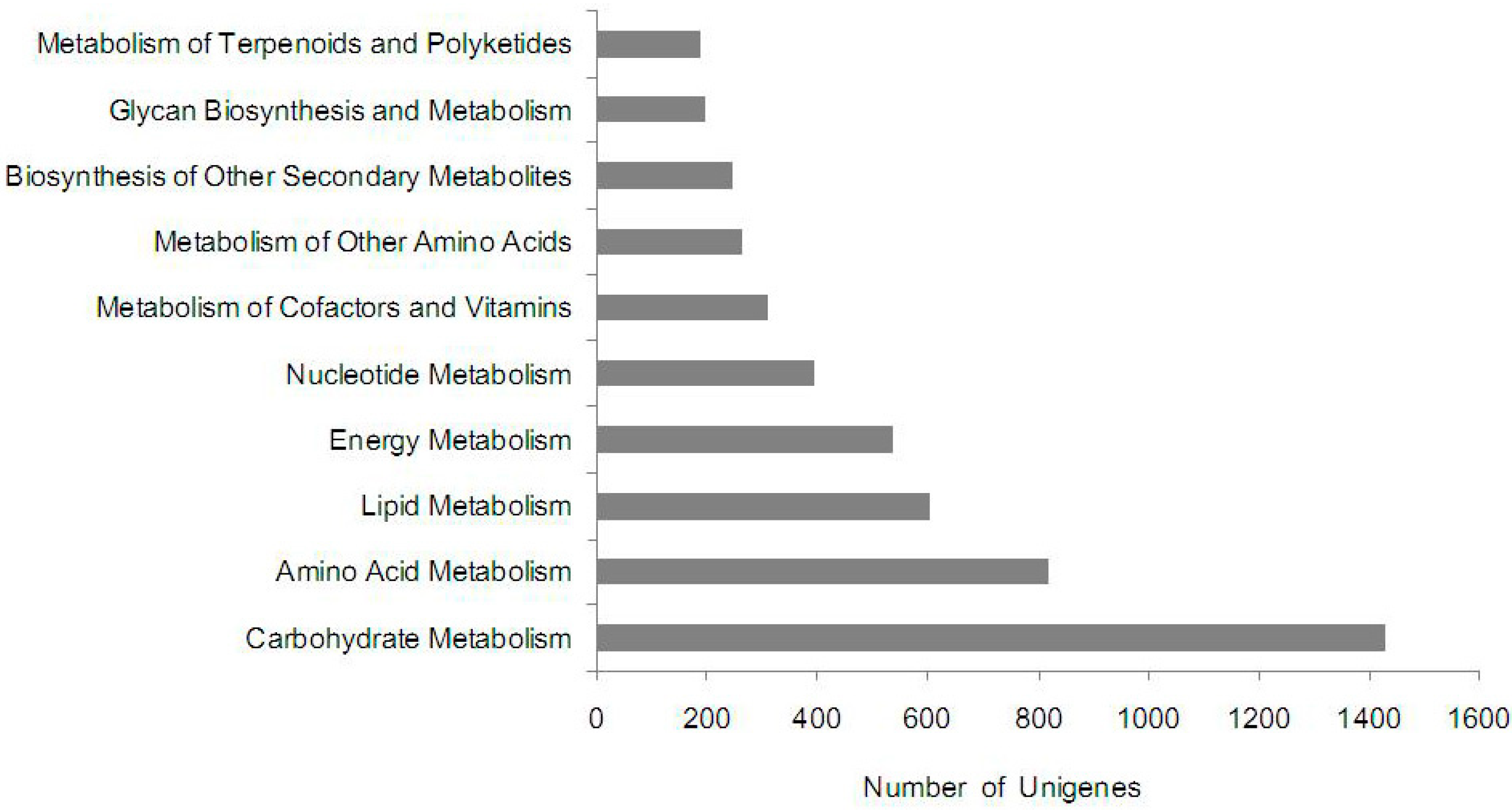
2.6. Differentially Expressed Unigenes between Non-Cracking Fruit and Cracking Fruit Libraries
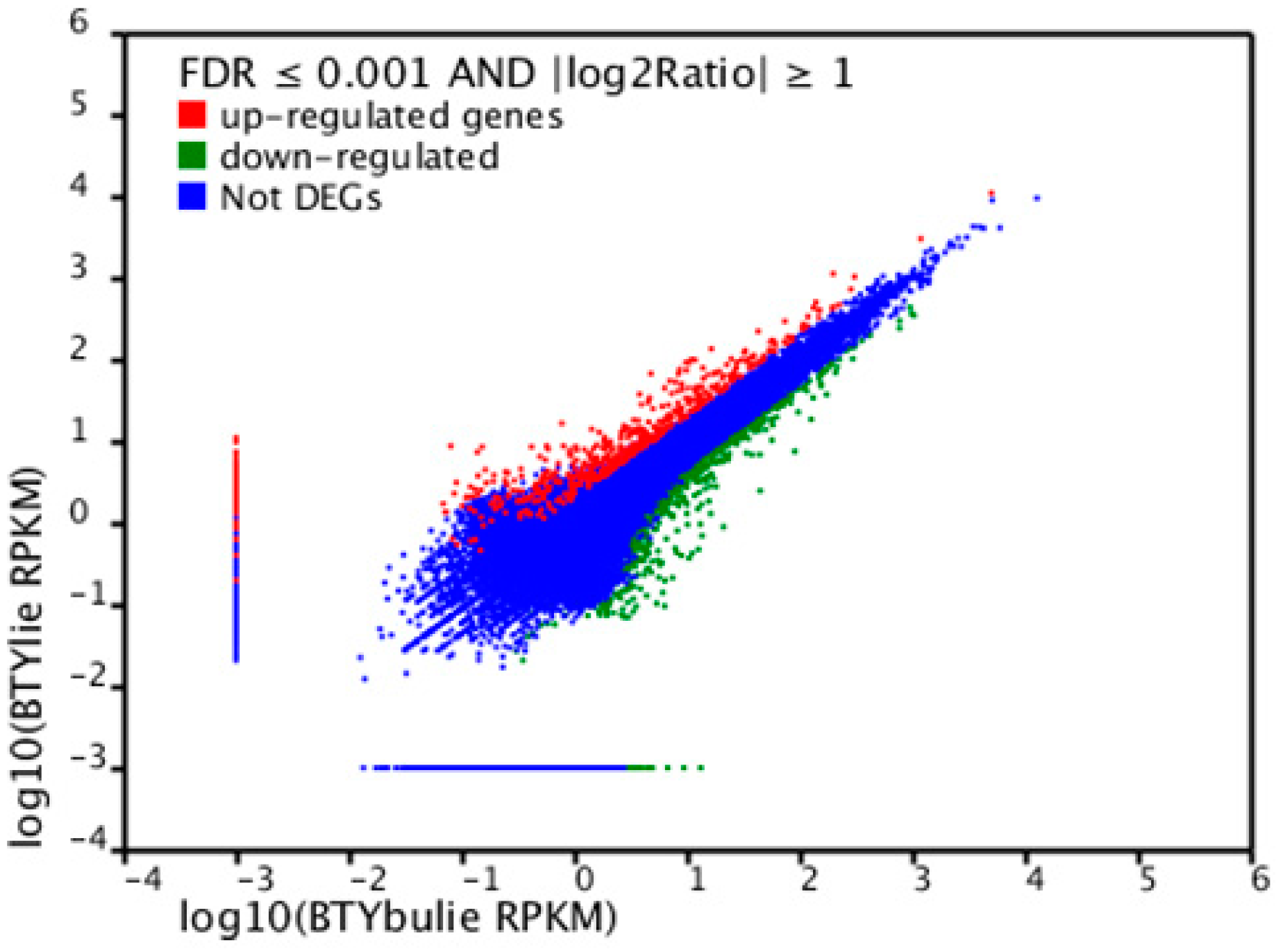
2.7. Identification of Candidate Genes for Fruit Cracking in Litchi chinensis Sonn.

3. Discussion
3.1. First Litchi Cracking Pericarp Reference Transcriptome Generated by RNA-Seq
3.2. Identification of Candidate Genes for Fruit Cracking in Water Transport
3.3. Identification of GA Metabolism-Related Candidate Genes for Fruit Cracking
3.4. Identification of Candidate ABA Metabolism-Related Genes for Fruit Cracking
3.5. Identification of Candidate Ca Transport-Related Genes for Fruit Cracking
3.6. Identification of Candidate Cell Wall Metabolism Genes for Fruit Cracking
4. Experimental Sections
4.1. Plant Materials
4.2. RNA Isolation and Library Construction for Transcriptome Analysis
4.3. Transcriptome Sequencing, de Novo Assembly and Functional Annotation
4.4. Identification of Differentially Expressed Unigenes
5. Conclusions

Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflict of Interest
References
- Huang, X.; Wang, H.C.; Li, J.; Yin, J.; Yuan, W.; Lu, J.; Huang, H.B. An overview of calcium’s role in lychee fruit cracking. Acta Hortic. 2003, 665, 1026–1035. [Google Scholar]
- Hoda, A.; Khalil, S.H.A. Cracking and fruit quality of pomegranate (Punica granatum L.) as affected by pre-harvest sprays of some growth regulators and mineral nutrients. J. Hortic. Sci. Ornam. Plants 2013, 5, 71–76. [Google Scholar]
- Lu, P.L.; Lin, C.H. Physiology of fruit cracking in wax apple (Syzygium samarangense). J. Bot. Orient. J. Plant Sci. 2011, 8, 70–76. [Google Scholar]
- Michelle, K.; Bruce, L.; Ken, S.; Carlos, H.C. Fruit skin side cracking and ostiole-end splitting shorten postharvest life in fresh figs (Ficus carica L.), but are reduced by deficit irrigation. Postharvest Biol. Technol. 2013, 85, 154–161. [Google Scholar]
- Lichter, A.; Dvir, O.; Fallik, E.; Cohen, S.; Golan, R.; Shemer, Z.; Sagi, M. Cracking of cherry tomatoes in solution. Postharvest Biol. Technol. 2002, 26, 305–312. [Google Scholar] [CrossRef]
- Caroline, G.; Chadoeuf, J.; Gilles, V.; Michel, G.; Francxoise, L. Cuticular cracking on nectarine fruit surface: Spatial distribution and development in relation to irrigation and thinning. J. Am. Soc. Hortic. Sci. 2007, 132, 583–591. [Google Scholar]
- Cristián, B.; Héctor, A.; Richard, M.; Gerardo, T.; Miguel, E.; Carolina, T.; José, A.Y.; José, Q.G.; Juan, C.R.; Herman, S. Cracking in sweet cherries: A comprehensive review from a physiological, molecular, and genomic perspective. J. Chil. J. Agric. Res. 2013, 73, 66–72. [Google Scholar]
- Measham, P.F.; Gracie, A.J.; Wilson, S.J.; Bound, S.A. Vascular flow of water induces side cracking in sweet cherry (Prunus avium L.). Adv. Hortic. Sci. 2010, 24, 243–248. [Google Scholar]
- Simon, G. Review on rain induced fruit cracking of sweet cherries (Prunus avium L.), its causes and the possibilities of prevention. Int. J. Hortic. Sci. 2006, 12, 27–35. [Google Scholar]
- Haq, I. Evaluation of Fruit Quality and Amelioration of Fruit Cracking in Litchi (Litchi chinensis Sonn.) Cutlivars through Water and Nutrient Management. Ph.D. Thesis, Khyber Pakhtunkhwa Agricultural University Peshawar, Peshawar, Pakistan, 2011. [Google Scholar]
- Li, J.G.; Huang, H.B.; Yuan, R.C.; Gao, F.F. Litchi fruit cracking in relation to fruit growth and water uptake kinetics. J. South China Agric. Univ. 1992, 13, 129–135. [Google Scholar]
- Cline, J.A.; Trought, M. Effect of gibberellic acid on fruit cracking and quality of Bing and Sam sweet cherries. Can. J. Plant Sci. 2007, 1, 545–552. [Google Scholar] [CrossRef]
- Munish, A.; Kahlon, P.S.; Mahajan, B.V.C. Effect of exogenous application of growth regulators on fruit drop, cracking and quality of litchi (Litchi chinensis sonn.) CV. Dehradun. Agric. Sci. Digest 2003, 23, 191–194. [Google Scholar]
- Yilmaz, C.; Ozguven, A.I. Hormon physiology of preharvest fruit acking in pomegranate (Punica granatum). Acta Hortic. 2003, 5, 716–727. [Google Scholar]
- Sharma, S.B.; Dhillon, B.S. Endogenous level of gibberellins in relation to fruit cracking in litchi (Litchi chinensis Sonn.). J. Res. Punjab Agric. Univ. 1986, 23, 432–434. [Google Scholar]
- Astuti, Y.A. The Effect of Preharvest CaCl2 Application Frequency on the Quality and Storage of Tomato Fruit. PhD Thesis, Bogor Agricultural University, Bogor, Indonesia, 2002; pp. 498–510. [Google Scholar]
- Huang, X.; Wang, H.C.; Li, J.; Yuan, W.; Lu, J.; Huang, H.B.; Yin, J. An overview of calcium’s role in lychee fruit cracking. Acta Hortic. 2005, 665, 231–240. [Google Scholar]
- Lu, W.J.; Wang, Y.; Jiang, Y.M.; Li, J.G.; Li, J.; Duan, X.; Song, L. Differential expression of litchi XET genes in relation to fruit growth. Plant Physiol. Biochem. 2006, 44, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Li, J.G.; Huang, X.M.; Huang, H.B. Comparison of the activities of enzymes related to cell wall metabolism in pericarp between litchi cultivars susceptible and resistant to fruit cracking. J. Plant Physiol. Mol. Biol. 2003, 29, 141–146. [Google Scholar]
- Wang, Y.; Lu, W.J.; Li, J.G.; Jiang, Y.M. Differential expression of two expansin genes in developing fruit of cracking-susceptible and -resistant litchi cultivars. J. Am. Soc. Hortic. Sci. 2006, 131, 118–121. [Google Scholar]
- Li, C.Q.; Wang, Y.; Huang, X.M.; Li, J.; Wang, H.C.; Li, J.G. De novo assembly and characterization of fruit transcriptome in Litchi chinensis Sonn. and analysis of differentially regulated genes in fruit in response to shading. BMC Genomics 2013, 14, 552–568. [Google Scholar]
- Flexas, J.; Ribas-Carbo, M.; Hanson, D.T.; Bota, J.; Otto, B.; Cifre, J.; Mcdowell, N.; Medrano, H.; Kaldenhoff, R. Tobacco aquaporin NtAQP1 is involved in mesophyll conductance to CO2 in vivo. Plant J. 2006, 48, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Aharon, R.; Shahak, Y.; Wininger, S.; Bendov, R.; Kapulnik, Y.; Galili, G. Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell 2003, 15, 439–447. [Google Scholar] [PubMed]
- Murai-Hatano, M.; Kuwagata, T.; Sakurai, J.; Nonami, H.; Ahamed, A.; Nagasuga, K. Effect of low root temperature on hydraulic conductivity of rice plants and the possible role of aquaporins. Plant Cell Physiol. 2008, 49, 1294–1305. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Lian, H.L.; Su, W.A. Role of the aquaporin PIP1 subfamily in the chilling tolerance of rice. Plant Cell Physiol. 2009, 50, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, F.M.; Bizzell, C.M.; Lee, D.J.; Zeevaart, J.A.; Amasino, R.M. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 2003, 15, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Tohru, A.; Kohji, M.; Sun, T.P.; Camille, M. Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor gibberellin insensitive dwarf1. Plant Cell 2008, 20, 2447–2459. [Google Scholar] [CrossRef] [PubMed]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.J.; Masatoshi, N.; Yoshihito, S.; Isomaro, Y. Cloning and characterization of the abscisic acidspecific glucosyltransferase gene from adzuki bean seedlings. Plant Physiol. 2002, 129, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Gosti, F.; Beaudoin, N.; Serizet, C.; Webb, A.A.; Vartanian, N. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 1999, 11, 1897–910. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Furuichi, T.; Cunningham, K.W.; Muto, S. A putative two-pore channel AtTPC1 mediates Ca2+ flux in Arabidopsis leaf cells. Plant Cell Physiol. 2001, 42, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, T.; Akahori, T.; Ashikari, M.; Maeshima, M. Expression of the vacuolar Ca2+/H+ exchanger, OsCAX1a, in rice: Cell and age specificity of expression, and enhancement by Ca2+. Plant Cell Physiol. 2006, 47, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Perez-Prat, E.; Narasimhan, L.; Binzel, M.L.; Botella, M.A.; Chen, Z.; Valpuesta, V.; Bressan, R.A.; Hasegawa, P.M. Induction of a putative Ca2+-ATPase mRNA in NaCl-adapted cells. Plant Physiol. 1992, 100, 1471–1478. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.K.; Cheong, Y.H.; Kim, K.N.; Luan, S. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 2004, 16, 1912–1924. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Over-expression of a single Ca2+- dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000, 23, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Brummell, D.A.; Harpster, M.H. Cell wall metabolism in fruit softening and quality and its manipulation in transgenicplants. Plant Mol. Biol. 2001, 47, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Schuch, W.; Kanczler, J.; Robertson, D.; Hobson, G.; Tucker, G.; Grierson, D.; Bright, S.; Bird, C. Fruit quality characteristics of transgenic tomato fruit with altered polygalacturonase activity. HortScience 1991, 26, 1517–1520. [Google Scholar]
- Amita, M.; Smriti, K.; Prabodh, K.T. Ethylene induced cotton leaf abscission is associated with higher expression of cellulase (GhCel1) and increased activities of ethylene biosynthesis enzymes in abscission zone. Plant Physiol. Biochem. 2008, 46, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Balbontín, C.; Ayala, H.; Carrasco, J.; Avilés, D. Transcriptional analysis of genes involved in cherry fruit cracking. VII Reunión Anual de Biología Vegetal: Pucón, Chile, Abstract in press. 2012. [Google Scholar]
- Knoche, M.; Peschel, S.; Hinz, M.; Bukovac, M. Studies on water transport through the sweet cherry fruit surface: Conductance of the cuticle in relation to fruit development. Planta 2001, 213, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Moctezuma, E.; Smith, D.L.; Gross, K.C. Antisense suppression of a beta-galactosidase gene (TBG6) in tomato increases fruit cracking. J. Exp. Bot. 2003, 54, 2025–2033. [Google Scholar]
- Wu, J.Y.; Peng, G.; Li, C.Q.; Lu, W.J.; Wang, Z.H.; Li, J.G. A new rapid and effective method for RNA isolation from litchi tissues of fruitlet and abscission zone. Acta Hortic. Sin. 2011, 38, 1191–1196. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.-C.; Wu, J.-Y.; Zhang, H.-N.; Shi, S.-Y.; Liu, L.-Q.; Shu, B.; Liang, Q.-Z.; Xie, J.-H.; Wei, Y.-Z. De Novo Assembly and Characterization of Pericarp Transcriptome and Identification of Candidate Genes Mediating Fruit Cracking in Litchi chinensis Sonn. Int. J. Mol. Sci. 2014, 15, 17667-17685. https://doi.org/10.3390/ijms151017667
Li W-C, Wu J-Y, Zhang H-N, Shi S-Y, Liu L-Q, Shu B, Liang Q-Z, Xie J-H, Wei Y-Z. De Novo Assembly and Characterization of Pericarp Transcriptome and Identification of Candidate Genes Mediating Fruit Cracking in Litchi chinensis Sonn. International Journal of Molecular Sciences. 2014; 15(10):17667-17685. https://doi.org/10.3390/ijms151017667
Chicago/Turabian StyleLi, Wei-Cai, Jian-Yang Wu, Hong-Na Zhang, Sheng-You Shi, Li-Qin Liu, Bo Shu, Qing-Zhi Liang, Jiang-Hui Xie, and Yong-Zan Wei. 2014. "De Novo Assembly and Characterization of Pericarp Transcriptome and Identification of Candidate Genes Mediating Fruit Cracking in Litchi chinensis Sonn." International Journal of Molecular Sciences 15, no. 10: 17667-17685. https://doi.org/10.3390/ijms151017667





