The L10P Polymorphism and Serum Levels of Transforming Growth Factor β1 in Human Breast Cancer
Abstract
:1. Introduction
2. Results and Discussion
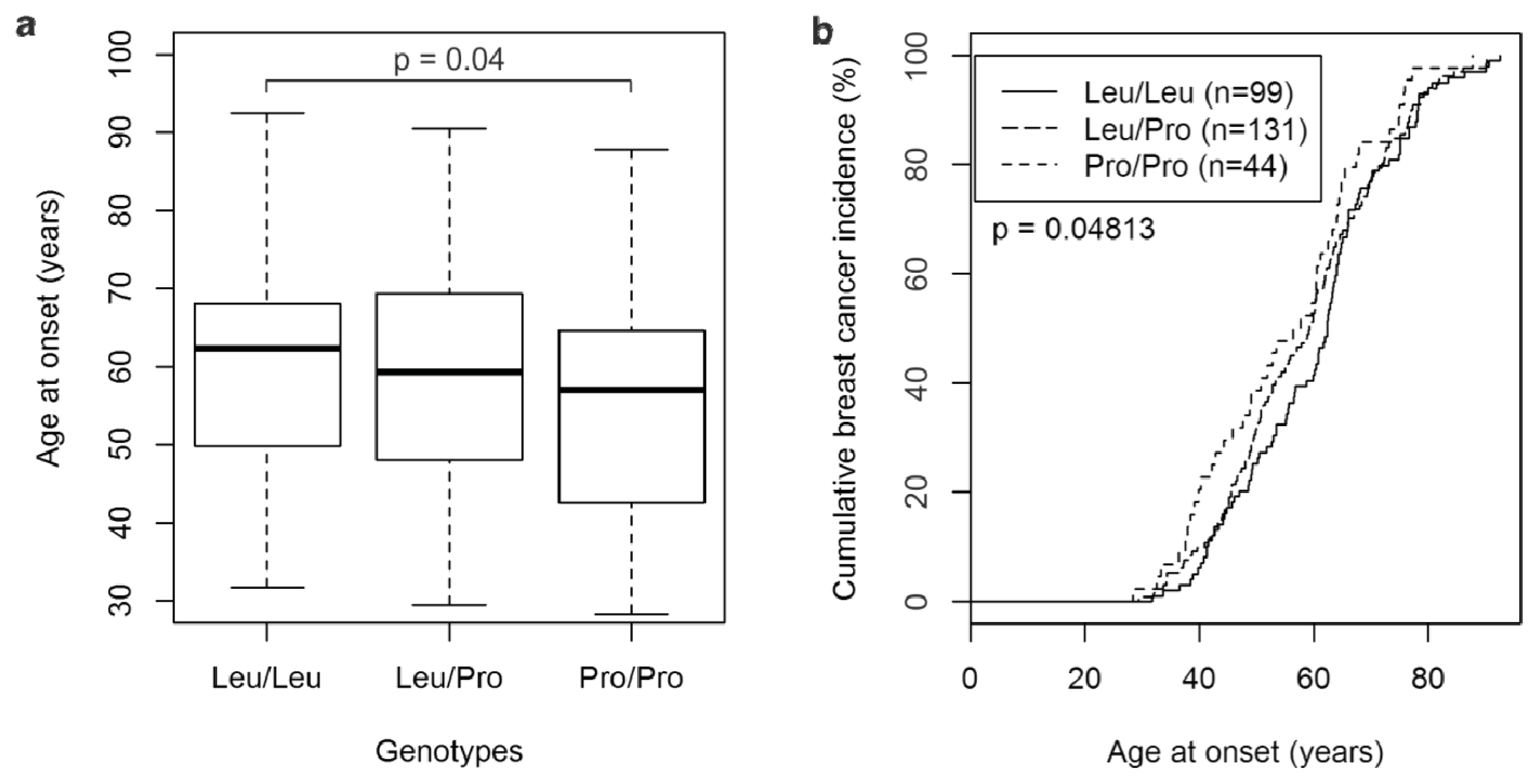
2.1. The TGFβ1 L10P SNP and Breast Cancer Risk
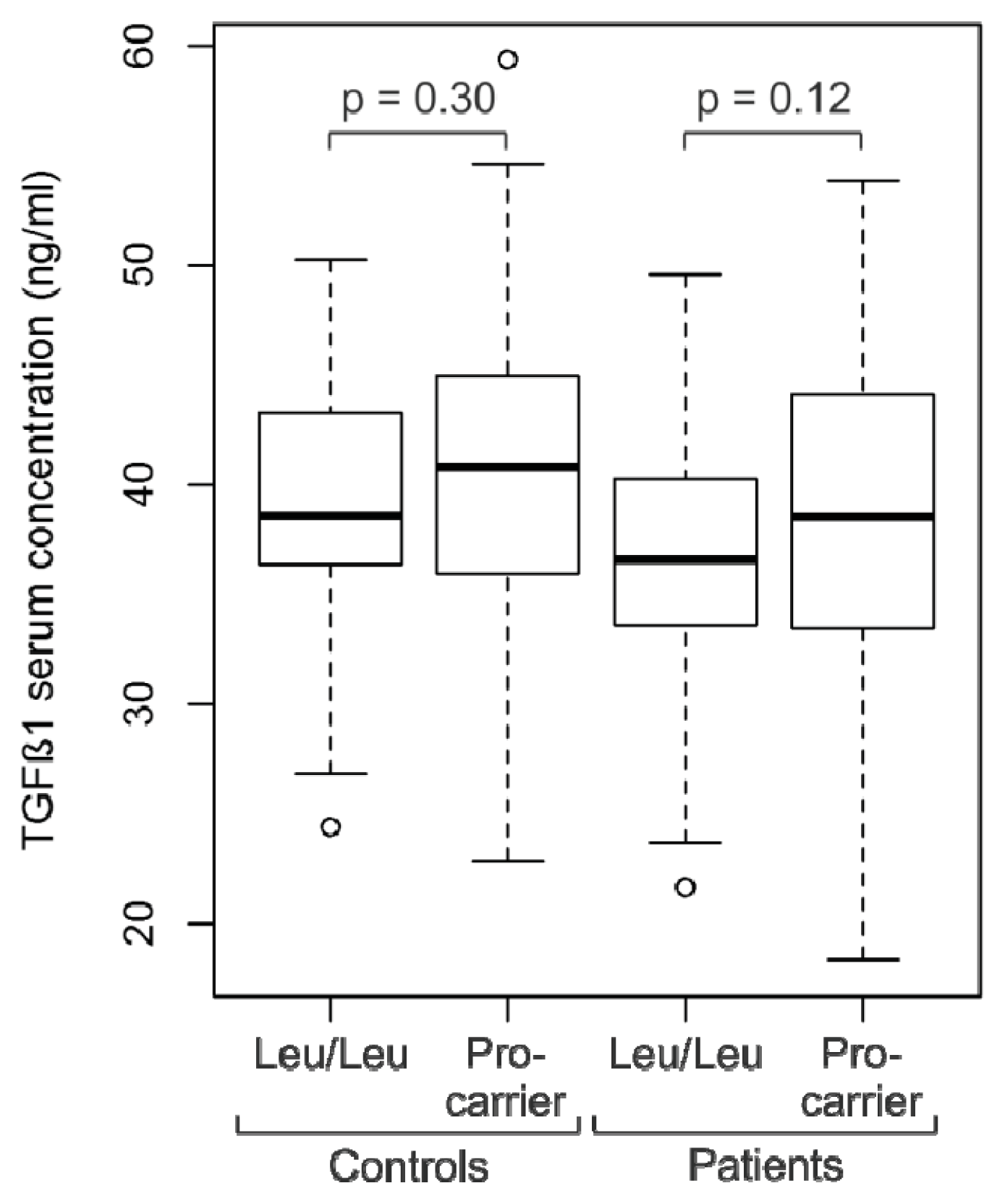
2.2. Analysis of TGFβ1 Serum Levels
2.3. Discussion
3. Experimental Section
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Massague, J. TGFbeta in Cancer. Cell 2008, 134, 215–230. [Google Scholar]
- Pardali, K.; Moustakas, A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim. Biophys. Acta 2007, 1775, 21–62. [Google Scholar]
- Siegel, P.M.; Massague, J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat. Rev. Cancer 2003, 3, 807–821. [Google Scholar]
- Padua, D.; Massague, J. Roles of TGF-beta in metastasis. Cell Res 2009, 19, 89–102. [Google Scholar]
- Imamura, T.; Hikita, A.; Inoue, Y. The roles of TGF-beta signaling in carcinogenesis and breast cancer metastasis. Breast Cancer 2012, 19, 118–124. [Google Scholar]
- Cox, A.; Dunning, A.M.; Garcia-Closas, M.; Balasubramanian, S.; Reed, M.W.; Pooley, K.A.; Scollen, S.; Baynes, C.; Ponder, B.A.; Chanock, S.; et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet 2007, 39, 352–358, Int. J. Mol. Sci. 2013, 14 15384. [Google Scholar]
- Dunning, A.M.; Ellis, P.D.; McBride, S.; Kirschenlohr, H.L.; Healey, C.S.; Kemp, P.R.; Luben, R.N.; Chang-Claude, J.; Mannermaa, A.; Kataja, V.; et al. A transforming growth factor beta1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res 2003, 63, 2610–2615. [Google Scholar]
- Lee, K.M.; Park, S.K.; Hamajima, N.; Tajima, K.; Yoo, K.Y.; Shin, A.; Noh, D.Y.; Ahn, S.H.; Hirvonen, A.; Kang, D. Genetic polymorphisms of TGF-beta1 & TNF-beta and breast cancer risk. Breast Cancer Res. Treat 2005, 90, 149–155. [Google Scholar]
- Shu, X.O.; Gao, Y.T.; Cai, Q.; Pierce, L.; Cai, H.; Ruan, Z.X.; Yang, G.; Jin, F.; Zheng, W. Genetic polymorphisms in the TGF-beta 1 gene and breast cancer survival: A report from the Shanghai Breast Cancer Study. Cancer Res 2004, 64, 836–839. [Google Scholar]
- Ziv, E.; Cauley, J.; Morin, P.A.; Saiz, R.; Browner, W.S. Association between the T29→C polymorphism in the transforming growth factor β1 gene and breast cancer among elderly white women: The study of osteoporotic fractures. JAMA 2001, 285, 2859–2863. [Google Scholar]
- Cox, D.G.; Penney, K.; Guo, Q.; Hankinson, S.E.; Hunter, D.J. TGFB1 and TGFBR1 polymorphisms and breast cancer risk in the Nurses’ Health Study. BMC Cancer 2007, 7, 175. [Google Scholar]
- Feigelson, H.S.; Patel, A.V.; Diver, W.R.; Stevens, V.L.; Thun, M.J.; Calle, E.E. Transforming growth factor beta receptor type I and transforming growth factor beta1 polymorphisms are not associated with postmenopausal breast cancer. Cancer Epidemiol. Biomark. Prev 2006, 15, 1236–1237. [Google Scholar]
- Jin, Q.; Hemminki, K.; Grzybowska, E.; Klaes, R.; Soderberg, M.; Zientek, H.; Rogozinska-Szczepka, J.; Utracka-Hutka, B.; Pamula, J.; Pekala, W.; et al. Polymorphisms and haplotype structures in genes for transforming growth factor beta1 and its receptors in familial and unselected breast cancers. Int. J. Cancer 2004, 112, 94–99. [Google Scholar]
- Kaklamani, V.G.; Baddi, L.; Liu, J.; Rosman, D.; Phukan, S.; Bradley, C.; Hegarty, C.; McDaniel, B.; Rademaker, A.; Oddoux, C.; et al. Combined genetic assessment of transforming growth factor-beta signaling pathway variants may predict breast cancer risk. Cancer Res 2005, 65, 3454–3461. [Google Scholar]
- Krippl, P.; Langsenlehner, U.; Renner, W.; Yazdani-Biuki, B.; Wolf, G.; Wascher, T.C.; Paulweber, B.; Bahadori, B.; Samonigg, H. The L10P polymorphism of the transforming growth factor-beta 1 gene is not associated with breast cancer risk. Cancer Lett 2003, 201, 181–184. [Google Scholar]
- Le Marchand, L.; Haiman, C.A.; van den Berg, D.; Wilkens, L.R.; Kolonel, L.N.; Henderson, B.E. T29C polymorphism in the transforming growth factor beta1 gene and postmenopausal breast cancer risk: The Multiethnic Cohort Study. Cancer Epidemiol. Biomark. Prev 2004, 13, 412–415. [Google Scholar]
- Mu, L.; Katsaros, D.; Lu, L.; Preti, M.; Durando, A.; Arisio, R.; Yu, H. TGF-β1 genotype and phenotype in breast cancer and their associations with IGFs and patient survival. Br. J. Cancer 2008, 99, 1357–1363. [Google Scholar]
- Rebbeck, T.R.; Antoniou, A.C.; Llopis, T.C.; Nevanlinna, H.; Aittomaki, K.; Simard, J.; Spurdle, A.B.; Couch, F.J.; Pereira, L.H.; Greene, M.H.; et al. No association of TGFB1 L10P genotypes and breast cancer risk in BRCA1 and BRCA2 mutation carriers: A multi-center cohort study. Breast Cancer Res. Treat 2009, 115, 185–192. [Google Scholar]
- Shin, A.; Shu, X.O.; Cai, Q.; Gao, Y.T.; Zheng, W. Genetic polymorphisms of the transforming growth factor-beta1 gene and breast cancer risk: A possible dual role at different cancer stages. Cancer Epidemiol. Biomark. Prev 2005, 14, 1567–1570. [Google Scholar]
- Sigurdson, A.J.; Hauptmann, M.; Chatterjee, N.; Alexander, B.H.; Doody, M.M.; Rutter, J.L.; Struewing, J.P. Kin-cohort estimates for familial breast cancer risk in relation to variants in DNA base excision repair, BRCA1 interacting and growth factor genes. BMC Cancer 2004, 4, 9. [Google Scholar]
- Hishida, A.; Iwata, H.; Hamajima, N.; Matsuo, K.; Mizutani, M.; Iwase, T.; Miura, S.; Emi, N.; Hirose, K.; Tajima, K. Transforming growth factor B1 T29C polymorphism and breast cancer risk in Japanese women. Breast Cancer 2003, 10, 63–69. [Google Scholar]
- Zheng, W. Genetic polymorphisms in the transforming growth factor-beta signaling pathways and breast cancer risk and survival. Methods Mol. Biol 2009, 472, 265–277. [Google Scholar]
- Guo, J.; Meng, H.; Pei, J.; Zhu, M. Association between the TNF-alpha-238G > A and TGF-beta1 L10P polymorphisms and breast cancer risk: A meta-analysis. Breast Care 2011, 6, 126–129. [Google Scholar]
- Huang, Y.; Li, B.; Qian, J.; Xie, J.; Yu, L. TGF-beta1 29T/C polymorphism and breast cancer risk: A meta-analysis involving 25,996 subjects. Breast Cancer Res. Treat 2010, 123, 863–868. [Google Scholar]
- Yokota, M.; Ichihara, S.; Lin, T.L.; Nakashima, N.; Yamada, Y. Association of a T29→C polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to myocardial infarction in Japanese. Circulation 2000, 101, 2783–2787. [Google Scholar]
- Li, X.; Yue, Z.C.; Zhang, Y.Y.; Bai, J.; Meng, X.N.; Geng, J.S.; Fu, S.B. Elevated serum level and gene polymorphisms of TGF-beta1 in gastric cancer. J. Clin. Lab Anal 2008, 22, 164–171. [Google Scholar]
- Proestling, K.; Hebar, A.; Pruckner, N.; Marton, E.; Vinatzer, U.; Schreiber, M. The pro allele of the p53 codon 72 polymorphism is associated with decreased intratumoral expression of BAX and p21, and increased breast cancer risk. PLoS One 2012, 7, e47325. [Google Scholar]
- R Development Core Team, R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria; p. 2009.
| Total | Leu/Leu | Leu/Pro | Pro/Pro | p-value | ||
|---|---|---|---|---|---|---|
| All subjects | 526 | 186 (35.4%) | 248 (47.1%) | 92 (17.5%) | ||
| Patients | 274 | 99 (36.1%) | 131 (47.8%) | 44 (16.1%) | 0.664 | |
| Controls | 252 | 87 (34.5%) | 117 (46.4%) | 48 (19.0%) | ||
| Patient subgroups | ||||||
| Tumor size | pT1 | 133 | 50 (37.6%) | 62 (46.6%) | 21 (15.8%) | 0.586 |
| pT2 | 55 | 20 (36.4%) | 27 (49.1%) | 8 (14.5%) | ||
| pT3, pT4 | 12 | 3 (25.0%) | 5 (41.7%) | 4 (33.3%) | ||
| other, na | 74 | 26 (35.1%) | 37 (50.0%) | 11 (14.9%) | ||
| Tumor type | ductal | 148 | 53 (35.8%) | 71 (48.0%) | 24 (16.2%) | 0.561 |
| lobular | 48 | 20 (41.7%) | 23 (47.9%) | 5 (10.4%) | ||
| other, na | 78 | 26 (33.3%) | 37 (47.4%) | 15 (19.2%) | ||
| Stage | I | 97 | 38 (39.2%) | 40 (41.2%) | 19 (19.6%) | 0.290 |
| II | 63 | 22 (34.9%) | 35 (55.6%) | 6 (9.5%) | ||
| III | 18 | 5 (27.8%) | 10 (55.6%) | 3 (16.7%) | ||
| other, na | 96 | 34 (35.4%) | 46 (47.9%) | 16 (16.7%) | ||
| Grade | pG1 | 42 | 17 (40.5%) | 21 (50.0%) | 4 (9.5%) | 0.405 |
| pG2 | 115 | 38 (33.0%) | 58 (50.4%) | 19 (16.5%) | ||
| pG3 | 88 | 36 (40.9%) | 35 (39.8%) | 17 (19.3%) | ||
| na | 29 | 8 (27.6%) | 17 (58.6%) | 4 (13.8%) | ||
| Lymph node status | pN0 | 143 | 56 (39.3%) | 62 (43.4%) | 25 (17.5%) | 0.320 |
| pN+ | 53 | 18 (34.0%) | 29 (54.7%) | 6 (11.3%) | ||
| na | 78 | 25 (32.1%) | 40 (51.3%) | 13 (16.7%) | ||
| ER status | pos | 201 | 75 (37.3%) | 100 (49.8%) | 26 (12.9%) | 0.085 |
| neg | 61 | 21 (34.4%) | 25 (41.0%) | 15 (24.6%) | ||
| na | 12 | 3 (25.0%) | 6 (50.0%) | 3 (25.0%) | ||
| PR status | pos | 138 | 47 (34.1%) | 73 (52.9%) | 18 (13.0%) | 0.130 |
| neg | 117 | 46 (39.3%) | 48 (41.0%) | 23 (19.7%) | ||
| na | 19 | 6 (31.6%) | 10 (52.6%) | 3 (15.8%) | ||
| HER2 status | pos | 51 | 24 (47.1%) | 21 (41.2%) | 6 (11.8%) | 0.176 |
| neg | 201 | 67 (33.3%) | 99 (49.3%) | 35 (17.4%) | ||
| na | 22 | 8 (36.4%) | 11 (50.0%) | 3 (13.6%) | ||
| p53 status | pos | 57 | 23 (40.4%) | 22 (38.6%) | 12 (21.1%) | 0.296 |
| neg | 190 | 68 (35.8%) | 94 (49.5%) | 28 (14.7%) | ||
| na | 27 | 8 (29.6%) | 15 (55.6%) | 4 (14.8%) | ||
| Genotypes/Alleles | OR | 95% c.i. | p-Value |
|---|---|---|---|
| Pro/Pro vs. Leu/Leu | 0.81 | 0.49–1.33 | 0.409 |
| Pro/Pro vs. Leu/Pro | 0.82 | 0.51–1.32 | 0.429 |
| Leu/Pro vs. Leu/Leu | 0.98 | 0.67–1.44 | 0.934 |
| Pro/Pro + Leu/Pro vs. Leu/Leu | 0.93 | 0.65–1.33 | 0.700 |
| Pro/Pro vs. Leu/Pro + Leu/Leu | 0.81 | 0.52–1.28 | 0.368 |
| Pro vs. Leu | 0.91 | 0.71–1.16 | 0.455 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Taubenschuß, E.; Marton, E.; Mogg, M.; Frech, B.; Ehart, L.; Muin, D.; Schreiber, M. The L10P Polymorphism and Serum Levels of Transforming Growth Factor β1 in Human Breast Cancer. Int. J. Mol. Sci. 2013, 14, 15376-15385. https://doi.org/10.3390/ijms140815376
Taubenschuß E, Marton E, Mogg M, Frech B, Ehart L, Muin D, Schreiber M. The L10P Polymorphism and Serum Levels of Transforming Growth Factor β1 in Human Breast Cancer. International Journal of Molecular Sciences. 2013; 14(8):15376-15385. https://doi.org/10.3390/ijms140815376
Chicago/Turabian StyleTaubenschuß, Eva, Erika Marton, Maurice Mogg, Barbara Frech, Lisa Ehart, Dana Muin, and Martin Schreiber. 2013. "The L10P Polymorphism and Serum Levels of Transforming Growth Factor β1 in Human Breast Cancer" International Journal of Molecular Sciences 14, no. 8: 15376-15385. https://doi.org/10.3390/ijms140815376





