Identification and Phylogenetic Analysis of a CC-NBS-LRR Encoding Gene Assigned on Chromosome 7B of Wheat
Abstract
:1. Introduction
2. Results
2.1. Amplification and Cloning of TdRGA-7Ba from Italy 363
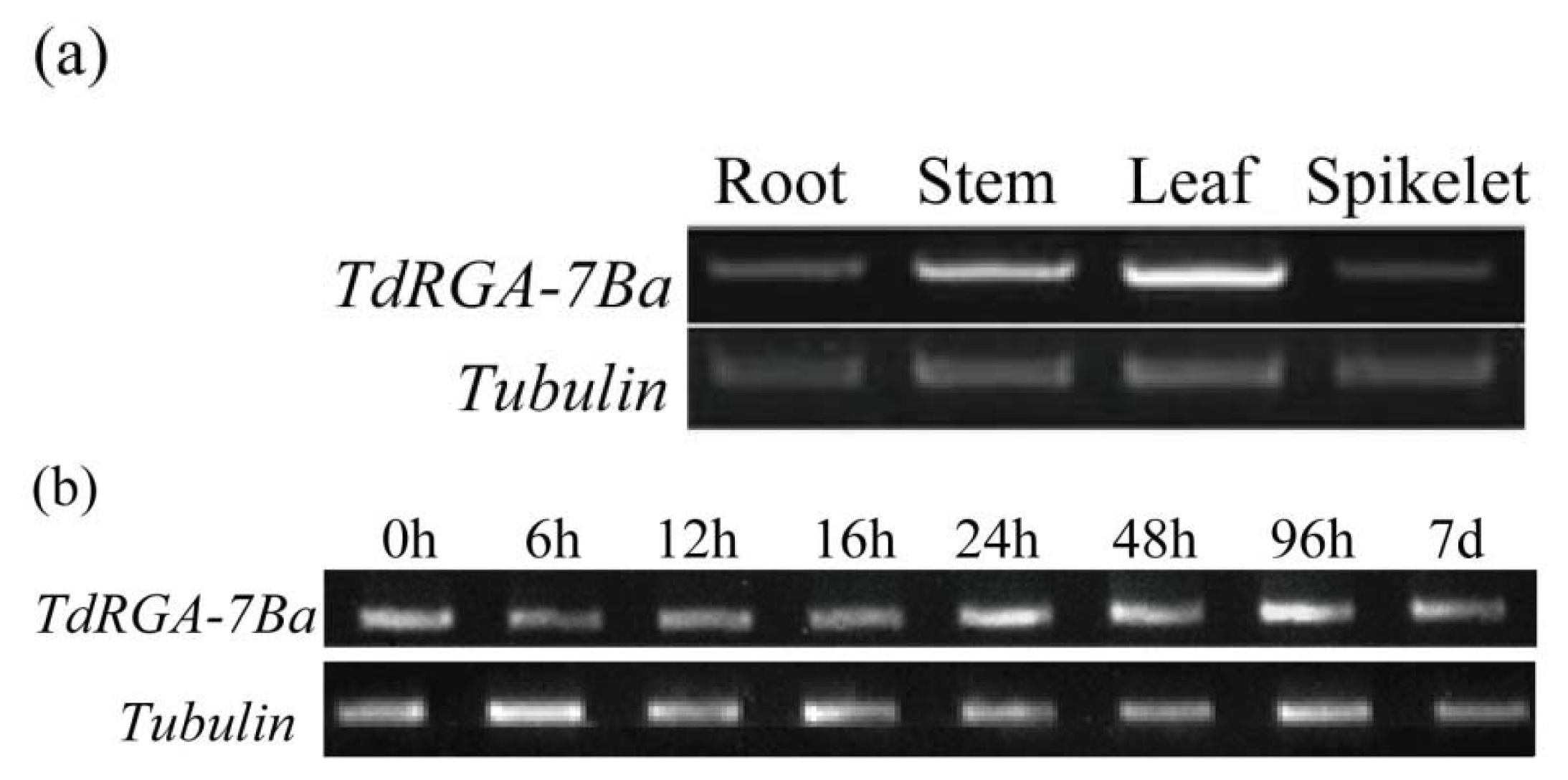
2.2. Expression Analysis of the TdRGA-7Ba
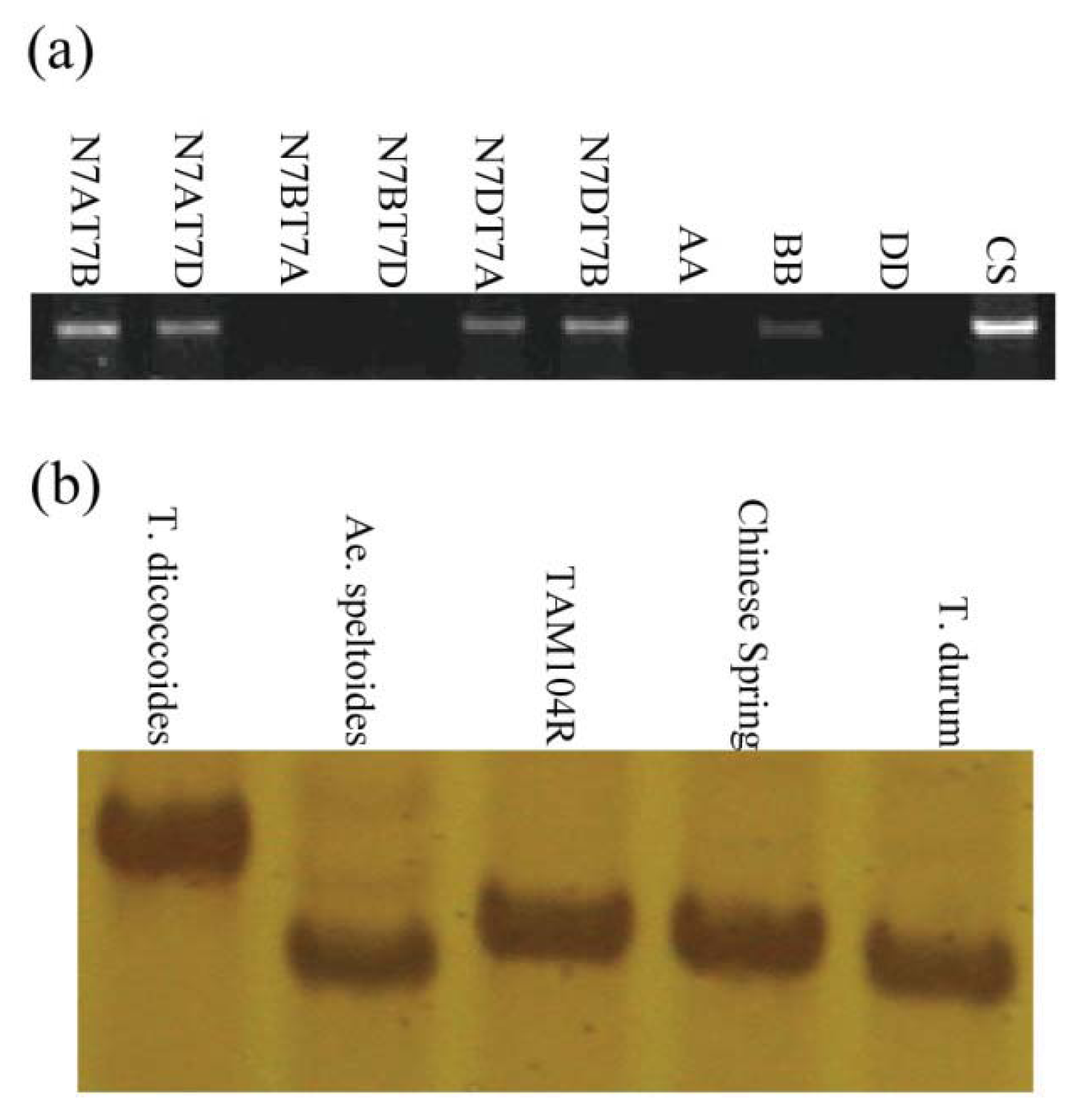
2.3. Chromosomal Assignment of TdRGA-7B Gene
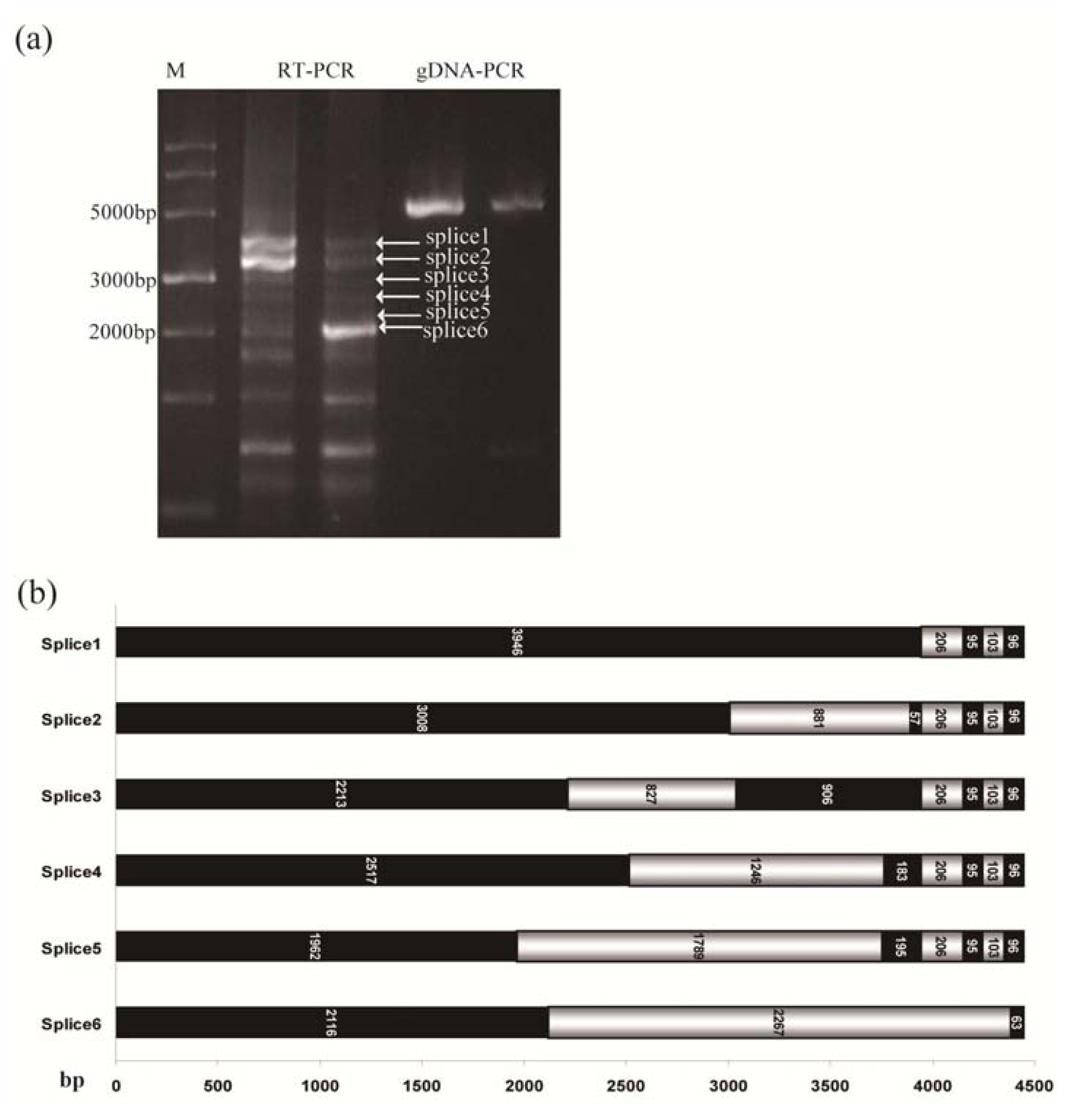
2.4. Alternative Splicing of TdRGA-7Ba
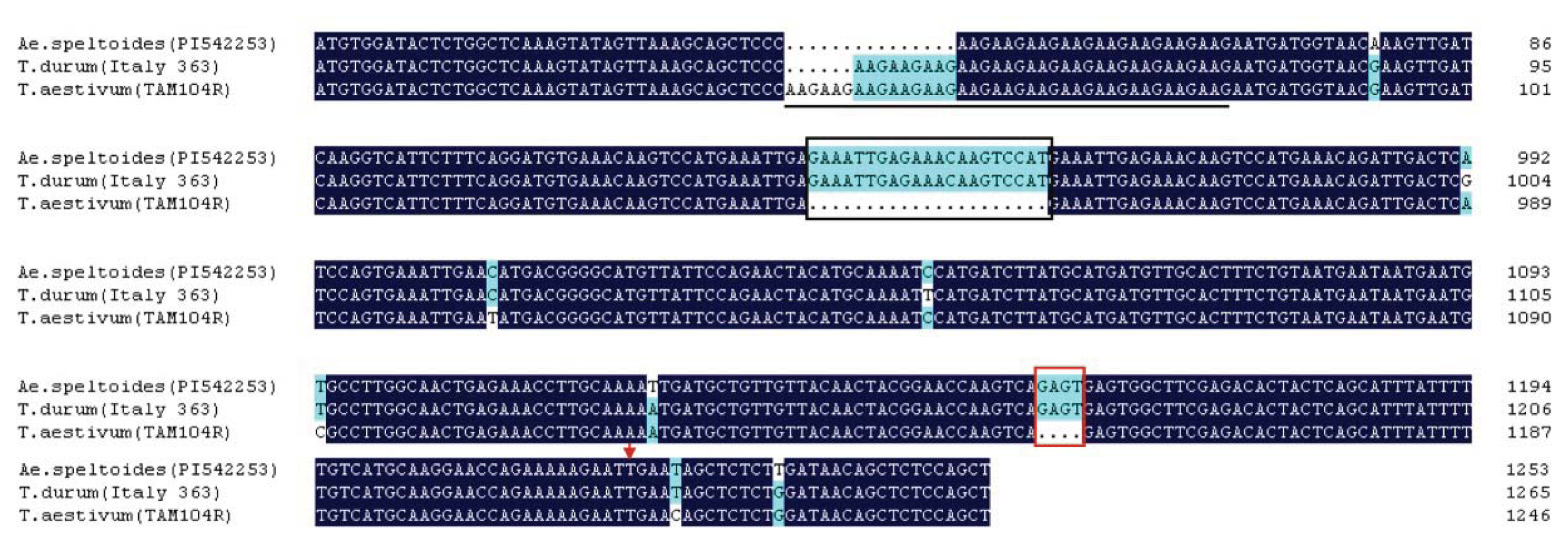
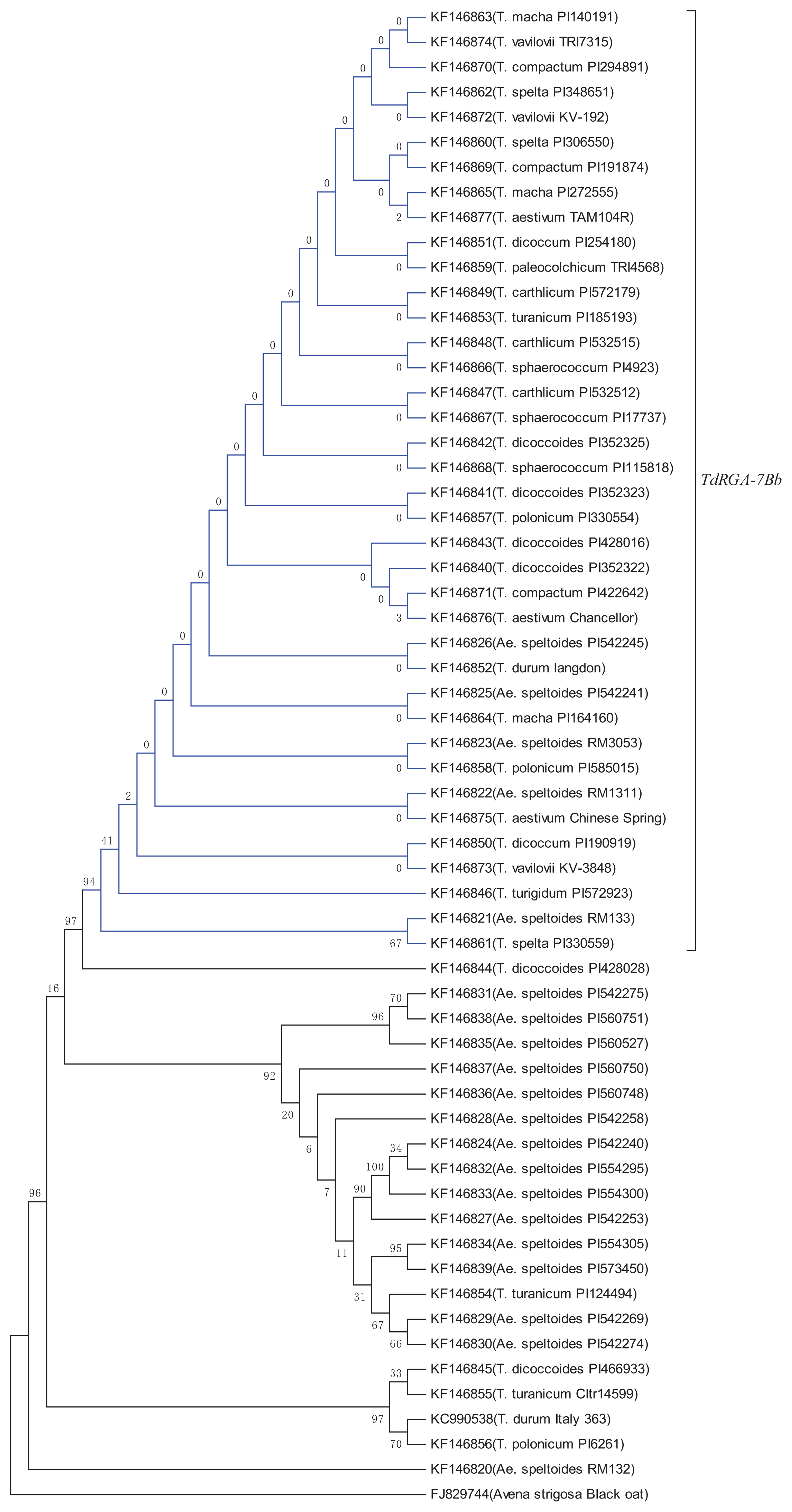
2.5. Genetic Variation and Phylogenetic Analysis of TdRGA-7B
2.6. The Development SSR Molecular Marker for TdRGA-7B
3. Discussion
4. Experimental Section
4.1. Plant Materials T. durum
4.2. DNA, RNA Extraction and cDNA Synthesis
4.3. TdRGA-7Ba Sequencing and Analysis
4.4. The Expression Pattern of TdRGA-7Ba
4.5. Chromosomal Assignment of TdRGA-7B Gene Sequence
4.6. Phylogenetic Analysis of TdRGA-7B
4.7. The Development of the SSR Molecular Marker for TdRGA-7B
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar]
- Bai, J.; Pennill, L.A.; Ning, J.; Lee, S.W.; Ramalingam, J.; Webb, C.A.; Zhao, B.; Sun, Q.; Nelson, J.C.; Leach, J.E. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res 2002, 12, 1871–1884. [Google Scholar]
- Tarr, D.E.; Alexander, H. TIR-NBS-LRR genes are rare in monocots: Evidence from diverse monocot orders. BMC Res. Notes 2009, 2, 197. [Google Scholar]
- Tameling, W.I.; Elzinga, S.D.; Darmin, P.S.; Vossen, J.H.; Takken, F.L.; Haring, M.A.; Cornelissen, B.J. The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 2002, 14, 2929–2939. [Google Scholar]
- Jiang, H.; Wang, C.; Ping, L.; Tian, D.; Yang, S. Pattern of LRR nucleotide variation in plant resistance genes. Plant Sci 2007, 173, 253–261. [Google Scholar]
- Tan, X.; Meyers, B.C.; Kozik, A.; West, M.A.; Morgante, M.; St Clair, D.A.; Bent, A.F.; Michelmore, R.W. Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC plant Biol 2007, 7, 56. [Google Scholar]
- Van der Hoorn, R.A.; de Wit, P.J.; Joosten, M.H. Balancing selection favors guarding resistance proteins. Trends Plant Sci 2002, 7, 67–71. [Google Scholar]
- Joshi, R.; Nayak, S. Functional characterization and signal transduction ability of nucleotide-binding site-leucine-rich repeat resistance genes in plants. Genet. Mol. Res 2011, 10, 2637–2652. [Google Scholar]
- Grant, J.J.; Chini, A.; Basu, D.; Loake, G.J. Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol. Plant Microbe Interact 2003, 16, 669–680. [Google Scholar]
- Kato, H.; Shida, T.; Komeda, Y.; Saito, T.; Kato, A. Overexpression of the activated disease resistance 1-like1 (ADR1-L1) gene results in a dwarf phenotype and activation of defense-related gene expression in Arabidopsis thaliana. J. Plant Biol 2011, 54, 172–179. [Google Scholar]
- Huang, L.; Brooks, S.; Li, W.; Fellers, J.; Nelson, J.C.; Gill, B. Evolution of new disease specificity at a simple resistance locus in a crop-weed complex: Reconstitution of the Lr21 gene in wheat. Genetics 2009, 182, 595–602. [Google Scholar]
- Meyers, B.C.; Kaushik, S.; Nandety, R.S. Evolving disease resistance genes. Curr. Opin. Plant Biol 2005, 8, 129–134. [Google Scholar]
- Ferrier-Cana, E.; Macadre, C.; Sevignac, M.; David, P.; Langin, T.; Geffroy, V. Distinct post-transcriptional modifications result into seven alternative transcripts of the CC-NBS-LRR gene JA1tr of Phaseolus vulgaris. Theor. Appl. Genet 2005, 110, 895–905. [Google Scholar]
- Zhang, X.-C.; Gassmann, W. RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell 2003, 15, 2333–2342. [Google Scholar]
- Gupta, P.; Mir, R.; Mohan, A.; Kumar, J. Wheat genomics: Present status and future prospects. Int. J. Plant Genomics 2008, 2008, 896451. [Google Scholar]
- Matsuoka, Y. Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol 2011, 52, 750–764. [Google Scholar]
- McFadden, E.; Sears, E. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered 1946, 37, 81–89. [Google Scholar]
- Huang, S.; Sirikhachornkit, A.; Su, X.; Faris, J.; Gill, B.; Haselkorn, R.; Gornicki, P. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 2002, 99, 8133–8138. [Google Scholar]
- Feldman, M.; Levy, A.A. Genome evolution due to allopolyploidization in wheat. Genetics 2012, 192, 763–774. [Google Scholar]
- Yue, J.X.; Meyers, B.C.; Chen, J.Q.; Tian, D.; Yang, S. Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol 2012, 193, 1049–1063. [Google Scholar]
- Kim, T.-H.; Kunz, H.-H.; Bhattacharjee, S.; Hauser, F.; Park, J.; Engineer, C.; Liu, A.; Ha, T.; Parker, J.E.; Gassmann, W.; et al. Natural variation in small molecule-induced TIR-NB-LRR signaling induces root growth arrest via EDS1- and PAD4-complexed R protein VICTR in Arabidopsis. Plant Cell 2012, 24, 5177–5192. [Google Scholar]
- Okuyama, Y.; Kanzaki, H.; Abe, A.; Yoshida, K.; Tamiru, M.; Saitoh, H.; Fujibe, T.; Matsumura, H.; Shenton, M.; Galam, D.C. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J 2011, 66, 467–479. [Google Scholar]
- De Majnik, J.; Ogbonnaya, F.C.; Moullet, O.; Lagudah, E.S. The Cre1 and Cre3 nematode resistance genes are located at homeologous loci in the wheat genome. Mol. Plant Microbe Interact 2003, 16, 1129–1134. [Google Scholar]
- Feuillet, C.; Travella, S.; Stein, N.; Albar, L.; Nublat, A.; Keller, B. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. USA 2003, 100, 15253–15258. [Google Scholar]
- Yahiaoui, N.; Srichumpa, P.; Dudler, R.; Keller, B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 2004, 37, 528–538. [Google Scholar]
- Law, C.; Wolfe, M. Location of genetic factors for mildew resistance and ear emergence time on chromosome 7B of wheat. Can. J. Genet. Cytol 1966, 8, 462–470. [Google Scholar]
- Hsam, S.; Huang, X.; Zeller, F. Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.) 6. Alleles at the Pm5 locus. Theor. Appl. Genet 2001, 102, 127–133. [Google Scholar]
- Huang, X.; Wang, L.; Xu, M.; Röder, M. Microsatellite mapping of the powdery mildew resistance gene Pm5e in common wheat (Triticum aestivum L.). Theor. Appl. Genet 2003, 106, 858–865. [Google Scholar]
- Mohler, V.; Bauer, A.; Bauer, C.; Flath, K.; Schweizer, G.; Hartl, L. Genetic analysis of powdery mildew resistance in German winter wheat cultivar Cortez. Plant Breed 2011, 130, 35–40. [Google Scholar]
- Xiao, M.; Song, F.; Jiao, J.; Wang, X.; Xu, H.; Li, H. Identification of the gene Pm47 on chromosome 7BS conferring resistance to powdery mildew in the Chinese wheat landrace Hongyanglazi. Theor. Appl. Genet 2013, 126, 1397–1403. [Google Scholar]
- Lin, F.; Xu, S.C.; Zhang, L.J.; Miao, Q.; Zhai, Q.; Li, L. SSR marker of wheat stripe rust resistance gene Yr2. J. Tritical Crops 2005, 25, 17–19. [Google Scholar]
- Li, Y.; Niu, Y.C. Identification of molecular markers for wheat stripe rust resistance gene Yr6. Acta Agr. Boreali-Sinica 2007, 22, 189–192. [Google Scholar]
- McIntosh, R.; Luig, N.; Baker, E. Genetic and cytogenetic studies of stem rust, leaf rust, and powdery mildew resistances in Hope and related wheat cultivars. Aust. J. Biol. Sci 1967, 20, 1181–1192. [Google Scholar]
- Sammeth, M.; Foissac, S.; Guigó, R. A general definition and nomenclature for alternative splicing events. PLoS Comput. Biol 2008, 4, e1000147. [Google Scholar]
- Johnson, J.M.; Castle, J.; Garrett-Engele, P.; Kan, Z.; Loerch, P.M.; Armour, C.D.; Santos, R.; Schadt, E.E.; Stoughton, R.; Shoemaker, D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 2003, 302, 2141–2144. [Google Scholar]
- Simpson, C.G.; Fuller, J.; Maronova, M.; Kalyna, M.; Davidson, D.; McNicol, J.; Barta, A.; Brown, J.W. Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J 2008, 53, 1035–1048. [Google Scholar]
- Kanazin, V.; Marek, L.F.; Shoemaker, R.C. Resistance gene analogs are conserved and clustered in soybean. Proc. Natl. Acad. Sci. USA 1996, 93, 11746–11750. [Google Scholar]
- Shen, K.A.; Meyers, B.C.; Islam-Faridi, M.N.; Chin, D.B.; Stelly, D.M.; Michelmore, R.W. Resistance gene candidates identified by PCR with degenerate oligonucleotide primers map to clusters of resistance genes in lettuce. Mol. Plant Microbe Interact 1998, 11, 815–823. [Google Scholar]
- Leister, D.; Kurth, J.; Laurie, D.A.; Yano, M.; Sasaki, T.; Devos, K.; Graner, A.; Schulze-Lefert, P. Rapid reorganization of resistance gene homologues in cereal genomes. Proc. Natl. Acad. Sci. USA 1998, 95, 370–375. [Google Scholar]
- Noir, S.; Combes, M.-C.; Anthony, F.; Lashermes, P. Origin, diversity and evolution of NBS-type disease-resistance gene homologues in coffee trees (Coffea L.). Mol. Genet. Genomics 2001, 265, 654–662. [Google Scholar]
- Gedil, M.A.; Slabaugh, M.B.; Berry, S.; Johnson, R.; Michelmore, R.; Miller, J.; Gulya, T.; Knapp, S.J. Candidate disease resistance genes in sunflower cloned using conserved nucleotide-binding site motifs: Genetic mapping and linkage to the downy mildew resistance gene Pl1. Genome 2001, 44, 205–212. [Google Scholar]
- Zamora, M.M.; Castagnaro, A.; Ricci, J.D. Isolation and diversity analysis of resistance gene analogues (RGAs) from cultivated and wild strawberries. Mol. Genet. Genomics 2004, 272, 480–487. [Google Scholar]
- Nair, R.A.; Thomas, G. Evaluation of resistance gene (R-gene) specific primer sets and characterization of resistance gene candidates in ginger (Zingiber officinale Rosc.). Curr. Sci 2007, 93, 61–66. [Google Scholar]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem 2010, 399, 257–261. [Google Scholar]
- Jordan, T.; Seeholzer, S.; Schwizer, S.; Töller, A.; Somssich, I.E.; Keller, B. The wheat Mla homologue TmMla1 exhibits an evolutionarily conserved function against powdery mildew in both wheat and barley. Plant J 2011, 65, 610–621. [Google Scholar]
- Huang, S.; Sirikhachornkit, A.; Faris, J.D.; Su, X.; Gill, B.S.; Haselkorn, R.; Gornicki, P. Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol. Biol 2002, 48, 805–820. [Google Scholar]
- Yamamoto, E.; Takashi, T.; Morinaka, Y.; Lin, S.; Wu, J.; Matsumoto, T.; Kitano, H.; Matsuoka, M.; Ashikari, M. Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice. Mol. Genet. Genomics 2010, 283, 305–315. [Google Scholar]
- Bomblies, K.; Lempe, J.; Epple, P.; Warthmann, N.; Lanz, C.; Dangl, J.L.; Weigel, D. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol 2007, 5, e236. [Google Scholar]
- Tian, D.; Traw, M.; Chen, J.; Kreitman, M.; Bergelson, J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 2003, 423, 74–77. [Google Scholar]
- Allen, G.; Flores-Vergara, M.; Krasynanski, S.; Kumar, S.; Thompson, W. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc 2006, 1, 2320–2325. [Google Scholar]

| Species | Source designation | Genome | Genotype | GenBank accession No. |
|---|---|---|---|---|
| Ae. speltoides | RM132 | BB | TdRGA-7Ba | KF146820 |
| Ae. speltoides | RM133 | BB | TdRGA-7Bb | KF146821 |
| Ae. speltoides | RM1311 | BB | TdRGA-7Bb | KF146822 |
| Ae. speltoides | RM3053 | BB | TdRGA-7Bb | KF146823 |
| Ae. speltoides var ligustica | PI542240 | BB | TdRGA-7Ba | KF146824 |
| Ae. speltoides var ligustica | PI542241 | BB | TdRGA-7Bb | KF146825 |
| Ae. speltoides | PI542245 | BB | TdRGA-7Bb | KF146826 |
| Ae. speltoides var. ligustica | PI542253 | BB | TdRGA-7Ba | KF146827 |
| Ae. speltoides | PI542258 | BB | TdRGA-7Ba | KF146828 |
| Ae. speltoides | PI542269 | BB | TdRGA-7Ba | KF146829 |
| Ae. speltoides | PI542274 | BB | TdRGA-7Ba | KF146830 |
| Ae. speltoides | PI542275 | BB | TdRGA-7Ba | KF146831 |
| Ae. speltoides | PI554295 | BB | TdRGA-7Ba | KF146832 |
| Ae. speltoides | PI554300 | BB | TdRGA-7Ba | KF146833 |
| Ae. speltoides var ligustica | PI554305 | BB | TdRGA-7Ba | KF146834 |
| Ae. speltoides var ligustica | PI560527 | BB | TdRGA-7Ba | KF146835 |
| Ae. speltoides | PI560748 | BB | TdRGA-7Ba | KF146836 |
| Ae. speltoides | PI560750 | BB | TdRGA-7Ba | KF146837 |
| Ae. speltoides | PI560751 | BB | TdRGA-7Ba | KF146838 |
| Ae. speltoides | PI573450 | BB | TdRGA-7Ba | KF146839 |
| T. dicoccoides | PI352322 | AABB | TdRGA-7Bb | KF146840 |
| T. dicoccoides | PI352323 | AABB | TdRGA-7Bb | KF146841 |
| T. dicoccoides | PI352325 | AABB | TdRGA-7Bb | KF146842 |
| T. dicoccoides | PI428016 | AABB | TdRGA-7Bb | KF146843 |
| T. dicoccoides | PI428028 | AABB | TdRGA-7Ba | KF146844 |
| T. dicoccoides | PI466933 | AABB | TdRGA-7Ba | KF146845 |
| T. turigidum | PI572923 | AABB | TdRGA-7Bb | KF146846 |
| T. carthlicum | PI532512 | AABB | TdRGA-7Bb | KF146847 |
| T. carthlicum | PI532515 | AABB | TdRGA-7Bb | KF146848 |
| T. carthlicum | PI572179 | AABB | TdRGA-7Bb | KF146849 |
| T. dicoccum | PI190919 | AABB | TdRGA-7Bb | KF146850 |
| T. dicoccum | PI254180 | AABB | TdRGA-7Bb | KF146851 |
| T. durum | Langdon | AABB | TdRGA-7Bb | KF146852 |
| T. durum | Italy 363 | AABB | TdRGA-7Ba | KC990538 |
| T. turanicum | PI185193 | AABB | TdRGA-7Bb | KF146853 |
| T. turanicum | PI124494 | AABB | TdRGA-7Ba | KF146854 |
| T. turanicum | CItr14599 | AABB | TdRGA-7Ba | KF146855 |
| T. polonicum | PI6261 | AABB | TdRGA-7Ba | KF146856 |
| T. polonicum | PI330554 | AABB | TdRGA-7Bb | KF146857 |
| T. polonicum | PI585015 | AABB | TdRGA-7Bb | KF146858 |
| T. paleocolchicum | TRI4568 | AABB | TdRGA-7Bb | KF146859 |
| T. spelta | PI306550 | AABBDD | TdRGA-7Bb | KF146860 |
| T. spelta | PI330559 | AABBDD | TdRGA-7Bb | KF146861 |
| T. spelta | PI348651 | AABBDD | TdRGA-7Bb | KF146862 |
| T. macha | PI140191 | AABBDD | TdRGA-7Bb | KF146863 |
| T. macha | PI164160 | AABBDD | TdRGA-7Bb | KF146864 |
| T. macha | PI272555 | AABBDD | TdRGA-7Bb | KF146865 |
| T. sphaerococcum | PI4923 | AABBDD | TdRGA-7Bb | KF146866 |
| T. sphaerococcum | PI17737 | AABBDD | TdRGA-7Bb | KF146867 |
| T. sphaerococcum | PI115818 | AABBDD | TdRGA-7Bb | KF146868 |
| T. compactum | PI191874 | AABBDD | TdRGA-7Bb | KF146869 |
| T. compactum | PI294891 | AABBDD | TdRGA-7Bb | KF146870 |
| T. compactum | PI422642 | AABBDD | TdRGA-7Bb | KF146871 |
| T. vavilovii | KU192 | AABBDD | TdRGA-7Bb | KF146872 |
| T. vavilovii | KU3848 | AABBDD | TdRGA-7Bb | KF146873 |
| T. vavilovii | TRI7315 | AABBDD | TdRGA-7Bb | KF146874 |
| T. aestivum | Chinese Spring | AABBDD | TdRGA-7Bb | KF146875 |
| T. aestivum | Chancellor | AABBDD | TdRGA-7Bb | KF146876 |
| T. aestivum | TAM104R | AABBDD | TdRGA-7Bb | KF146877 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gong, C.; Cao, S.; Fan, R.; Wei, B.; Chen, G.; Wang, X.; Li, Y.; Zhang, X. Identification and Phylogenetic Analysis of a CC-NBS-LRR Encoding Gene Assigned on Chromosome 7B of Wheat. Int. J. Mol. Sci. 2013, 14, 15330-15347. https://doi.org/10.3390/ijms140815330
Gong C, Cao S, Fan R, Wei B, Chen G, Wang X, Li Y, Zhang X. Identification and Phylogenetic Analysis of a CC-NBS-LRR Encoding Gene Assigned on Chromosome 7B of Wheat. International Journal of Molecular Sciences. 2013; 14(8):15330-15347. https://doi.org/10.3390/ijms140815330
Chicago/Turabian StyleGong, Caiyan, Shuanghe Cao, Renchun Fan, Bo Wei, Guiping Chen, Xianping Wang, Yiwen Li, and Xiangqi Zhang. 2013. "Identification and Phylogenetic Analysis of a CC-NBS-LRR Encoding Gene Assigned on Chromosome 7B of Wheat" International Journal of Molecular Sciences 14, no. 8: 15330-15347. https://doi.org/10.3390/ijms140815330




