Abstract
In bacteria, glycogen or oligosaccharide accumulation involves glucose-1-phosphate partitioning into either ADP-glucose (ADP-Glc) or UDP-Glc. Their respective synthesis is catalyzed by allosterically regulated ADP-Glc pyrophosphorylase (EC 2.7.7.27, ADP-Glc PPase) or unregulated UDP-Glc PPase (EC 2.7.7.9). In this work, we characterized the UDP-Glc PPase from Streptococcus mutans. In addition, we constructed a chimeric protein by cutting the C-terminal domain of the ADP-Glc PPase from Escherichia coli and pasting it to the entire S. mutans UDP-Glc PPase. Both proteins were fully active as UDP-Glc PPases and their kinetic parameters were measured. The chimeric enzyme had a slightly higher affinity for substrates than the native S. mutans UDP-Glc PPase, but the maximal activity was four times lower. Interestingly, the chimeric protein was sensitive to regulation by pyruvate, 3-phosphoglyceric acid and fructose-1,6-bis-phosphate, which are known to be effectors of ADP-Glc PPases from different sources. The three compounds activated the chimeric enzyme up to three-fold, and increased the affinity for substrates. This chimeric protein is the first reported UDP-Glc PPase with allosteric regulatory properties. In addition, this is a pioneer work dealing with a chimeric enzyme constructed as a hybrid of two pyrophosphorylases with different specificity toward nucleoside-diphospho-glucose and our results turn to be relevant for a deeper understanding of the evolution of allosterism in this family of enzymes.
1. Introduction
The fate of Glc-1P in sugar anabolism involves a first step where the Glc moiety is “activated” by the formation of a nucleoside-diphospho-glucose (NDP-Glc) catalyzed by different NDP-Glc pyrophosphorylases (NDP-Glc PPases). Later, diverse glycosyl transferases with specificity toward a particular NDP-Glc lead the monosaccharide to a variety of carbohydrate metabolic routes. In general, in bacteria, there are two major biochemical roles for nucleotide-linked sugars: as intermediates in the formation of monosaccharides used in the production of complex carbohydrates, via UDP-Glc or as glycosyl donors for glycogen synthesis, using ADP-Glc [,]. These two key metabolites are products of either UDP-Glc or ADP-Glc PPase, through a reaction that requires a divalent metal ion (physiologically Mg2+): U(A)TP + Glc-1P ↔ U(A)DP-Glc + PPi. Other specific NDP-sugar PPases complement the metabolic scenario for the production of the multiple mono-, oligo-, and poly-saccharides in the cell, which are found as free components or covalently bound to proteins and lipids [,].
UDP-Glc PPase (EC 2.7.7.9) is ubiquitously distributed in all types of organisms, and it plays a critical role in carbohydrates metabolism []. Significant differences at the level of amino acids sequence and three-dimensional structure found between the enzymes from prokaryotes and eukaryotes imply that they are not homologous. Eukaryotic UDP-Glc PPases are bigger than those found in bacteria [,], and the enzyme from Entamoeba histolytica (and probably from all protozoa) was recently characterized as being regulated by redox modification of critical cysteinyl residues []. Many bacterial UDP-Glc PPases have been characterized [–], and the crystallographic structures of the enzyme from Escherichia coli [], Sphingomonas elodea [] and Corynebacterium glutamicum [] have been elucidated. The prokaryotic UDP-Glc PPase is a dimeric/tetrameric protein formed by a single subunit of ~35 kDa with a relatively high specific activity and specificity for Glc-1P and UTP [–].
A main characteristic among prokaryotic NDP-sugar PPases (including UDP-Glc PPase) is that they are non-regulated enzymes. However, ADP-Glc PPase (EC 2.7.7.27) is an exception in that it catalyzes the key regulatory step in the pathway for glycogen and starch biosynthesis in bacteria and plants, respectively. Most ADP-Glc PPases characterized so far are allosterically regulated by metabolites that are principal intermediates in the major carbon assimilation pathway in the respective organism [,]. The three-dimensional structure of the homotetrameric forms of the enzyme from potato tuber and Agrobacterium tumefaciens has been recently solved by X-ray crystallography [,]. Structural studies have determined that ADP-Glc PPases are larger than other prokaryotic NDP-sugar PPases. The former enzymes have an N-terminal catalytic domain (structurally similar to all PPases) that contains the active site, plus an additional C-terminal domain (absent in other PPases). On the basis of different studies [–], it has been proposed that the distinctive C-domain in ADP-Glc PPases is functionally related to allosteric regulation. Herein, we report the molecular cloning and heterologous expression of the gene coding for UDP-Glc PPase from Streptococcus mutans. Also, we constructed a chimeric protein by fusing to the latter S. mutans enzyme the C-terminal domain of the E. coli ADP-Glc PPase. The resulting hybrid protein retained UDP-Glc PPase activity and exhibited allosteric properties, being activated by 3-phosphoglycerate (3-PGA), fructose-1,6-bis-phosphate (Fru-1,6-bisP) and pyruvate (Pyr). Our results have an impact on understanding the structure-to-function relationship between domains in PPases as well as the strategic changes driven by evolution to awaken allosterism in proteins.
2. Results and Discussion
2.1. Isolation and Analysis of the Gene Coding for UDP-Glc PPase in S. mutans and Construction of the Chimeric Protein
The genome elucidated for S. mutans UA159 indicates the presence of a single gene coding for a putative UDP-Glc PPase (SMU_322c; Gene ID: 1029376) []; known as galU according to previous reports [,,,] or gtaB in other sources [,]. To determine the functional role of galU in S. mutans we amplified this single gene from S. mutans ATCC 25175 using specific primers properly designed (see details under Experimental Section) based on the database information available for S. mutans UA159 []. The amplified gene (the sequence of which was deposited in NCBI; GenBank accession number KC626324) codes for a protein 100% identical to the one in the genome of the reference strain. The S. mutans galU gene codes for a protein (SmuGalU) with a theoretical molecular mass of 33.9 kDa and a 33.8% and 40.4% identity with UDP-Glc PPases from C. glutamicum and Helicobacter pylori, respectively. The former was used to elucidate the active site geometry in this type of enzymes [] and the H. pylori protein for determining the enzymatic reaction mechanism (which was bi-bi ordered) []. Also, the protein coded by S. mutans galU shares similar identity to the UDP-Glc PPase from E. coli (41.5%); Sphingomonas elodea (41.1%) and Streptomyces coelicolor (43.8%), which have been structurally and kinetically characterized [,,].
The S. mutans GalU has a high identity (85.1%) with the UDP-Glc PPase from S. pneumoniae [,]. The gene amplified from S. mutans ATCC 25175 was utilized for two main purposes (Figure 1): (i) to insert it into the pRSET-B expression plasmid, looking to produce the recombinant UDP-Glc PPase (SmuGalU) as a tool for the structural and kinetic characterization; and (ii) to construct a gene coding for a chimeric protein, seeking to investigate the functionality of key domains in PPases. Amino acid sequence alignment between UDP-Glc and ADP-Glc PPases shows that these proteins share a homologous N-terminal domain, which is involved in catalysis. However, the enzymes specific for ADP-Glc are larger proteins with an extra C-terminal domain that is presumably related to allosteric properties as indicated above (Figure 1a, amplified in Figure S1). To advance in the latter hypothesis, we produced a hybrid protein by fusing the putative regulatory C-domain (from the E. coli ADP-Glc PPase) to the C-terminal of the non-allosteric UDP-Glc PPase from S. mutans (Figure 1b).
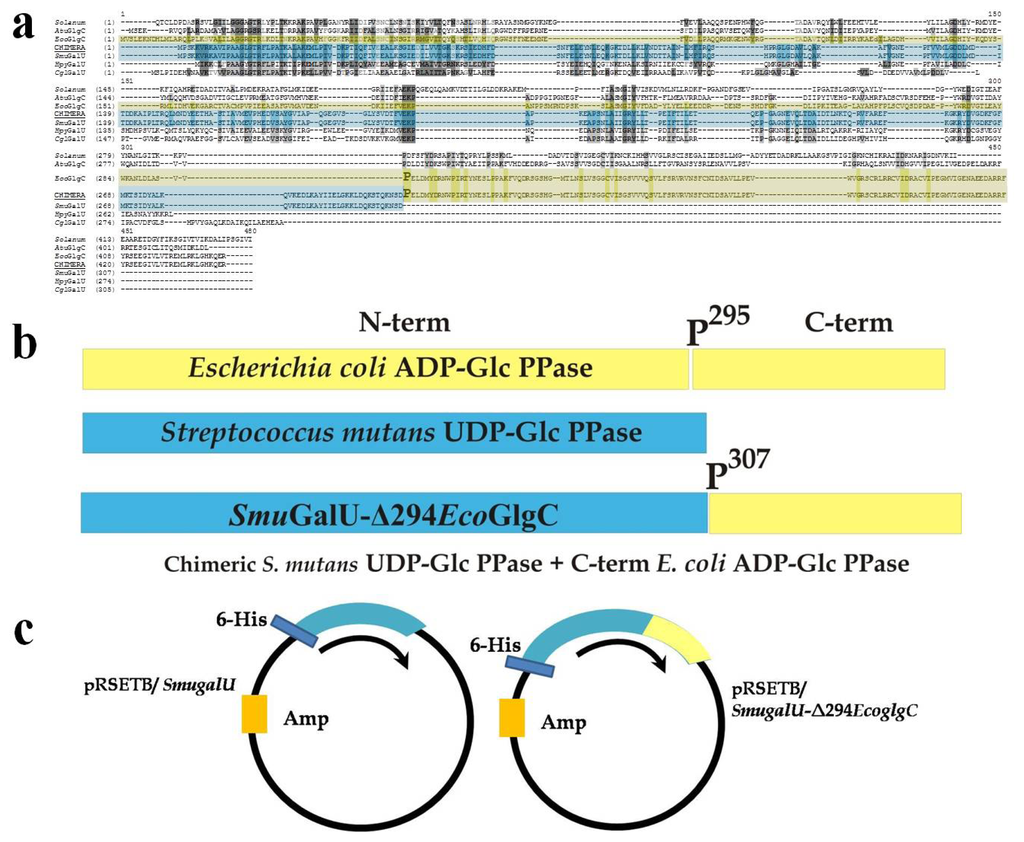
Figure 1.
Schematic representation for chimeric SmuGalU-Δ294EcoGlgC construction. (a) Alignment between different ADP-glucose pyrophosphorylases (ADP-Glc PPases) and UDP-Glc PPases (Ref: Solanum: ADP-Glc PPase small subunit from potato; AtuGlgC, A. tumefaciens ADP-Glc PPase; EcoGlgC, E. coli ADP-Glc PPase; Chimera, chimeric SmuGalU-Δ294EcoGlgC; SmuGalU, S. mutans UDP-Glc PPase; HpyGalU, H. pylory UDP-Glc PPase; CglGalU, C. glutamicum UDP-Glc PPase). E. coli ADP-Glc PPase P295 is over-marked; (b) Construction of chimeric enzyme: The C-terminus belonging to the ADP-Glc PPase from E. coli (yellow) was “added” to the entire UDP-Glc PPase from S. mutans ATCC 25175 (light-blue); (c) Plasmids used to express both proteins.
Figure 1 details the “cut and paste” strategy used to construct the chimeric gene coding for the hybrid protein SmuGalU-Δ294EcoGlgC. Hence, we pasted a DNA fragment cut from the E. coli glgC gene, which codes for the 137 C-terminal amino acidic residues of the ADP-Glc PPase (starting at codon belonging to P295 residue, which is in a connector loop) to the entire S. mutans galU gene. The resulting 1329 pb DNA piece was used to construct a pRSET-B derivative plasmid (Figure 1c) suitable to express a 443 amino acid chimeric protein with a theoretical molecular mass of 49.4 kDa. Previously chimeric enzymes were obtained and characterized after switching/swapping N- and C-terminal domains between ADP-Glc PPases (specifically involving the enzymes from E. coli and A. tumefaciens [] or from prokaryotic and eukaryotic photosynthetic organisms []). This is the first report regarding a hybrid UDP-Glc PPase/ADP-Glc PPase protein.
2.2. Structural and Kinetic Characterization of SmuGalU and Chimeric SmuGalU-Δ294EcoGlgC
Both SmuGalU and chimeric SmuGalU-Δ294EcoGlgC were produced as recombinant proteins via heterologous expression in E. coli using the respective pRSET-B derivative plasmid (see details in Figure 1). Thus, both proteins were obtained having a His-tag fused at the N-terminus. When E. coli BL21 (DE3) was used as a host, native SmuGalU was over-expressed in soluble fractions but SmuGalU-Δ294EcoGlgC was mostly recovered in the pellet (inclusion bodies). The chimeric protein appeared in soluble fractions when the expression host was turned to E. coli BL21 (DE3) pLysS. Culture conditions and procedures were similar for both hosts (see Experimental Section). After inducing expression and obtaining crude extracts from the transformed cells, the recombinant His-tagged proteins were purified by immobilized metal (Ni2+) affinity chromatography, after which they reached a high degree of purity according to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis (Figure 2a). Size exclusion chromatography on Superdex 200 revealed that under soluble conditions SmuGalU and SmuGalU-Δ294EcoGlgC arranged homotetrameric quaternary structures of a molecular mass of ~150 kDa and ~200 kDa, respectively (Figure 2b). Results obtained with SmuGalU are in good agreement with tetrameric structures previously determined for UDP-Glc PPases from different sources, e.g., both crystallized enzymes from E. coli and S. elodea [,]. On the other hand, the native form found for SmuGalU-Δ294EcoGlgC indicates that this chimeric protein shares similar oligomeric properties to the polypeptides that form its hybrid structure.
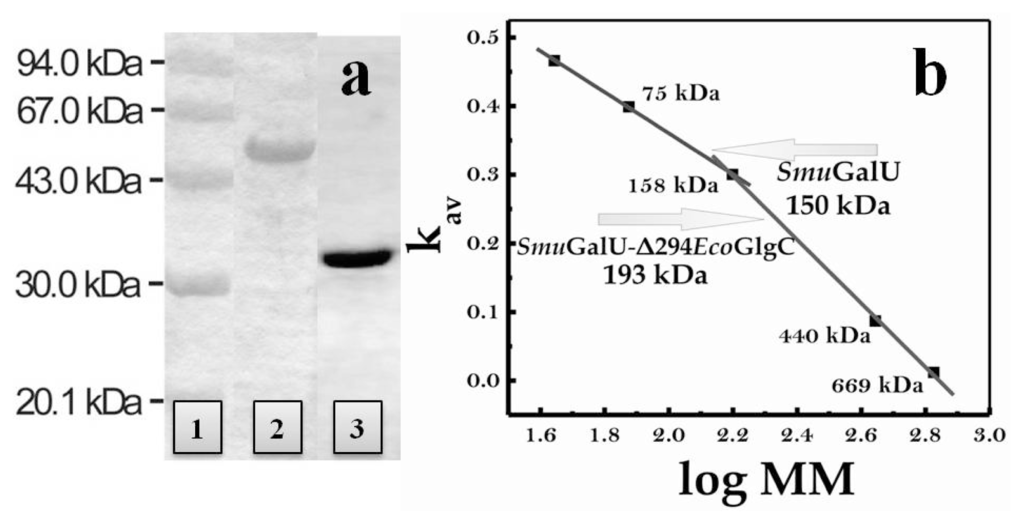
Figure 2.
(a) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of recombinant UDP-Glc PPase from S. mutans and chimeric SmuGalU-Δ294EcoGlgC after purification. Lane 1: Molecular weight markers; Lane 2: chimeric SmuGalU-Δ294EcoGlgC; Lane 3: His-tagged SmuGalU. Purifications were conducted as described in the Experimental Section; (b) Molecular mass determination, performed from size exclusion chromatography, as detailed in the Experimental Section.
Purified SmuGalU and SmuGalU-Δ294EcoGlgC had UDP-Glc PPase activity. It catalyzed the synthesis of UDP-Glc and PPi from Glc-1P and UTP (in the presence of 3 mM Mg2+) with specific activities of 40 and 11 U/mg, respectively. It is well known that a divalent metal ion (commonly Mg2+) is an essential cofactor for ADP-Glc PPases [,] and UDP-Glc PPases [,,,,]. In our hands, the activities of SmuGalU and SmuGalU-Δ294EcoGlgC were strictly dependent of Mg2+. Both proteins were fully active at ~3 mM and inhibited by higher concentrations of the divalent cation (Figure 3). Other metal ions could replace Mg2+, as illustrated by Figure 4. At 0.5 mM Mn2+SmuGalU reached a four-fold higher activity than with 3 mM Mg2+, but the enzyme was inhibited at higher Mn2+ levels. This enzyme was also active with Cd2+, Ca2+, Co2+, Ni2+, Cu2+ and Cr2+, with higher activity at 0.5 mM of the metal ion and inhibition at different levels with higher amounts of the respective divalent cofactor (Figure 4a). Concerning SmuGalU-Δ294EcoGlgC, the protein was as active as with 3 mM Mg2+ when assayed with 0.5 mM of Mn2+, Ca2+, Co2+, or Cu2+, whereas with 0.5 mM Cd2+, Ni2+, or Cr2+ it showed only about half the activity. Except for Mn2+ and Ca2+, the other divalent cations inhibited the hybrid protein at higher concentrations (Figure 4b).
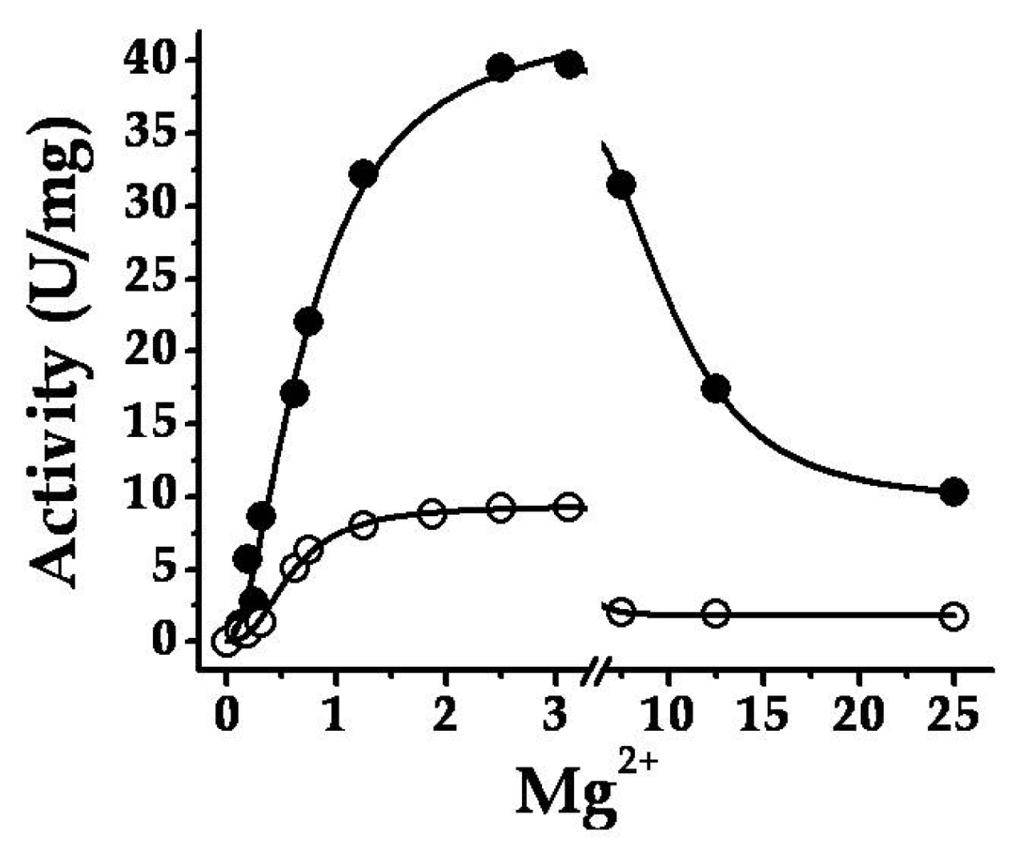
Figure 3.
Mg2+ curves for both SmuGalU (filled circles) and chimeric SmuGalU-Δ294EcoGlgC (empty circles).
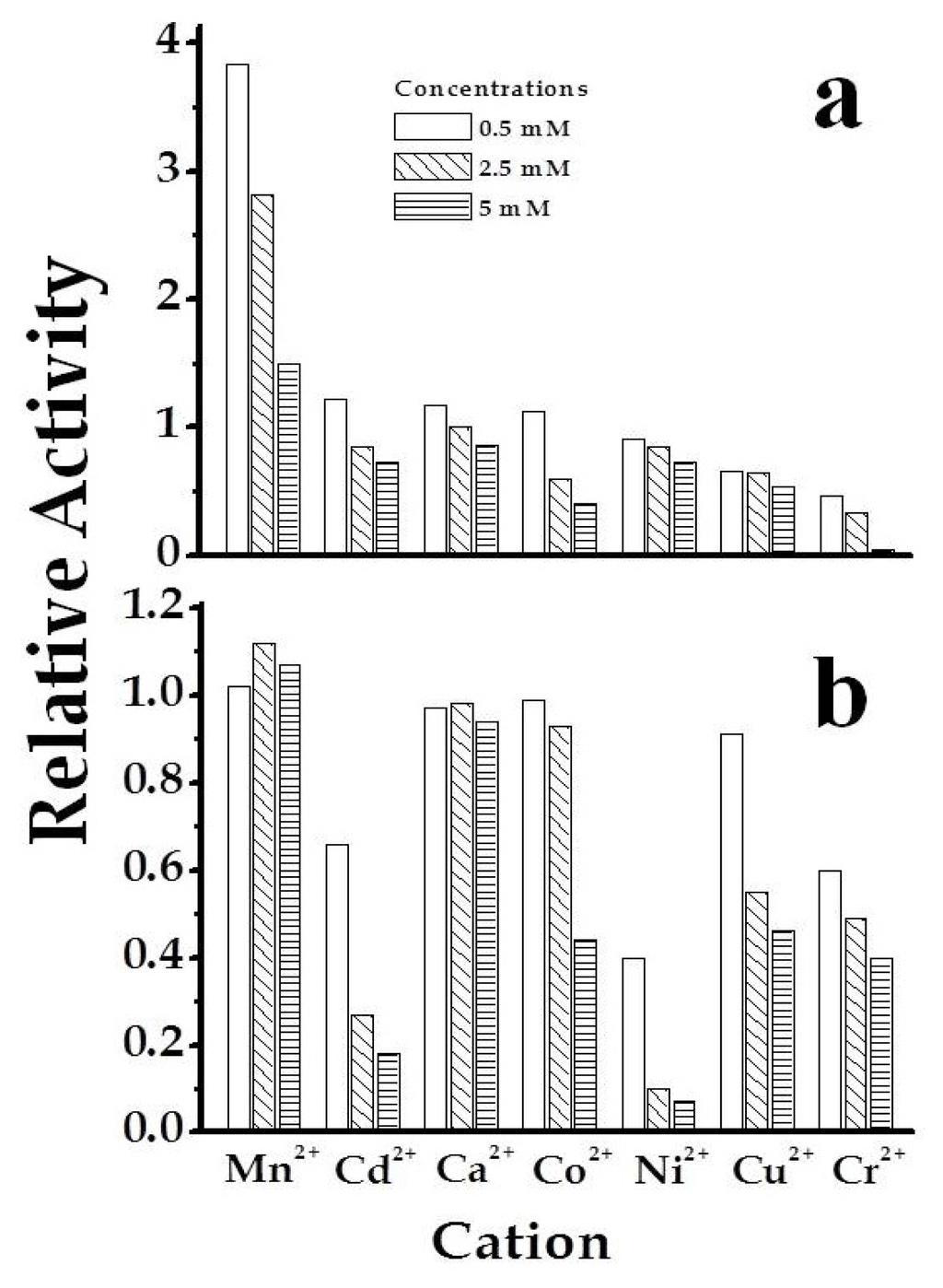
Figure 4.
Use of different divalent metal ions by SmuGalU (a) and chimeric SmuGalU-Δ294EcoGlgC (b). Empty bars correspond to 0.5 mM, sparse filled bars to 2.5 mM and dense filled bars to 5 mM of the corresponding metal analyzed. Controls are related to the enzyme activity measure in the same conditions, using 3 mM Mg2+.
A similar pH-dependence of activity was observed for SmuGalU and SmuGalU-Δ294EcoGlgC when studied in the range of pH 6.0–10.0. Both proteins exhibited a maximum at pH 8.0, and sharply decreased below pH 7.0 (data not shown). These results are in good agreement with those previously reported for pneumococcal UDP-Glc PPase, which is fully active at pH 8.0–8.5 [], as well as for ADP-Glc PPases from different sources, where optimal activity is around pH 8.0 [,]. After the behavior of the proteins in respect to pH and the requirement for divalent metal ions were studied, the further kinetic characterization was performed at pH 8.0 and 3 mM Mg2+ as its saturating concentration. The kinetic parameters of SmuGalU and SmuGalU-Δ294EcoGlgC for the substrates Glc-1P, UTP and divalent cofactor Mg2+ are summarized in Table 1. The analysis of these parameters is needed to compare the kinetic properties of these recombinant proteins and understand the functionality of the different structure domains in PPases.

Table 1.
Kinetic parameters for SmuGalU and SmuGalU-Δ294EcoGlgC. Parameters were calculated from averaged data from three independent experiments, as detailed in the Experimental Section.
SmuGalU slightly deviated from a hyperbolic behavior for both UTP and Glc-1P displaying positive cooperativity. Saturation curves for Mg2+ were even more sigmoidal (Table 1). Apparent affinities for the different substrates/cofactor exhibited by SmuGalU were in the same order of magnitude as those reported for other bacterial UDP-Glc PPases so far characterized [,,,,]. The recombinant enzyme reached a Vmax of 62 U/mg (Table 1), which was significantly higher than the UDP-Glc PPases from E. coli [], S. elodea [] and S. pneumoniae [], although not as high as UDP-Glc PPase from S. coelicolor [] and in the same order as the enzyme from Xanthomonas spp. []. Concerning SmuGalU-Δ294EcoGlgC, saturation curves also showed a slight or marked deviation from the hyperbolic behavior for the substrates or the divalent cation cofactor, respectively (Table 1). The chimeric protein increased three-fold in the apparent affinity for UTP regarding the UDP-Glc PPase from S. mutans, whilst affinities for Glc-1P and Mg2+ remained at the same level. Besides, the chimeric protein exhibited a four-fold lower Vmax when compared with SmuGalU (Table 1). Then, the results suggest that the C-terminal domain from the fusion of the E. coli ADP-Glc PPase to the S. mutans UDP-Glc PPase modifies the enzyme to acquire a conformation with slightly reduced ratio Vmax/S0.5 (analogous to ratio Vmax/Km, defined as catalytic efficiency for hyperbolic kinetics) (Table 1). The kinetic properties exhibited by SmuGalU-Δ294EcoGlgC are remarkable for a hybrid protein composed by domains of PPases with different specificity. Although our studies are the first dealing with kinetic characterization of an UDP-Glc PPase/ADP-Glc PPase chimeric enzyme, they can be compared with previous hybrids between ADP-Glc PPases. Thus, when N- and C-terminal domains from the E. coli and A. tumefaciens ADP-Glc PPases were switched, the chimeric construct exhibited activities between 20% and 30% of the value of the original wild-type enzymes [].
In general, prokaryotic NDP-sugar PPases are relatively specific for their substrates, although some exceptions have been reported. For example, dTTP is a substrate in some bacterial UDP-Glc PPases [,,], whereas GDP-mannose (GDP-Man) PPases from M. tuberculosis and from Leptospira interrogans exhibit promiscuity in the use of the nucleotide triphosphate (NTP) substrate [,]. Different NTPs (ATP; UTP; ITP; GTP; CTP; dTTP) at 0.1, 0.5 and 2.5 mM final concentration were assayed as alternative substrates for SmuGalU and SmuGalU-Δ294EcoGlgC. It was observed that S. mutans UDP-Glc PPase was active with dTTP as an alternative to UTP. The activity in the synthesis direction of dTDP-Glc was four-fold lower than the production of UDP-Glc, although the enzyme showed similar affinity for either NTP (S0.5 0.54 mM, at 1 mM Glc-1P and 3 mM Mg2+). Results with SmuGalU are in accordance with previous reports indicating that these enzymes (from a prokaryotic source) are capable of utilizing dTTP [,,]. Conversely, chimeric SmuGalU-Δ294EcoGlgC exhibited a high specificity toward UTP.
2.3. The Chimeric SmuGalU-Δ294EcoGlgC Protein Exhibits Allosteric Properties
Activation-inhibition assays were performed for both recombinant proteins under study (SmuGalU and SmuGalU-Δ294EcoGlgC) testing several compounds known to be important effectors of ADP-Glc PPases from different sources [,]. The metabolites utilized were phosphoenolpyruvate (PEP); fructose-1,6-bisphosphate (Fru-1,6-bisP), pyruvate (Pyr), 3-phosphoglyceric acid (3-PGA), Glc-6P, ribose-5P (Rib-5P), fructose-6P (Fru-6P), Man-1P, Man-6P, Pi, AMP, ADP, NAD+, NADH, NADP+ and NADPH. These effectors were tested at up to 10 mM (Pyr was even varied up to 150 mM) under activity assay conditions that were saturating and non-saturating in respect to the amount of substrates in the medium. None of the metabolites analyzed affected the activity of SmuGalU. This insensitivity to regulation is in good agreement with data reported for bacterial UDP-Glc PPases, which in general are not regulated by allosteric effectors []. On the other hand, Pyr, Fru-1,6-bisP and 3-PGA activated chimeric SmuGalU-Δ294EcoGlgC (Figure 5) (none of the molecules analyzed inhibited the enzyme). This fact per se is one of the striking results of this work, since it constitutes the first report regarding a protein having UDP-Glc PPase activity (or a PPase activity other than ADP-Glc PPase) and being subjected to allosteric regulation.
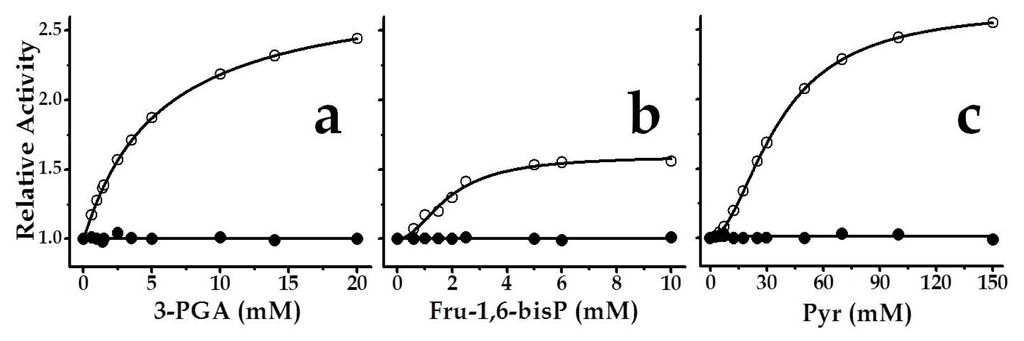
Figure 5.
Saturation curves for chimeric SmuGalU-Δ294EcoGlgC effectors: (a) 3-PGA, (b) Fru-1,6-bisP and (c) Pyr. Filled circles belong to SmuGalU and empty circles to chimeric SmuGalU-Δ294EcoGlgC enzyme. Reactions were conducted at 1 mM Glc-1P, 1 mM UTP and 3 mM Mg2+. Values of relative activity were calculated based on activities measured in the absence of effector, specifically 40 U/mg and 11 U/mg for SmuGalU and SmuGalU-Δ294EcoGlgC, respectively.
Saturation curves for each metabolite activating SmuGalU-Δ294EcoGlgC are detailed in Figure 5. Both, 3-PGA and Pyr, increased the Vmax of the hybrid fused protein by 2.4- and 2.6-fold, respectively, while Fru-1,6-bisP activation was 1.6-fold. Respective to the apparent capacity for binding of the allosteric effector, the chimeric enzyme behaved with the highest apparent affinity for Fru-1,6-bisP (A0.5 2.0 mM), and with positive cooperativity (nH 1.5). For 3-PGA the behavior was hyperbolic (nH 1.0) with an A0.5 value of 5.4 mM, whereas Pyr depicted a sigmoidal (nH 1.5) saturation curve from which an A0.5 of 30 mM could be calculated (Figure 5). It is worth noting that this apparent affinity value of the chimeric protein calculated for Pyr is in the same order as what was found for the interaction (also activating) of this metabolite with the E. coli ADP-Glc PPase []. 3-PGA and Fru-1,6-bisP are the main activators of plants and some bacterial ADP-Glc PPases [,] as well, although with higher affinities.
It was valuable to analyze the kinetic parameters of SmuGalU-Δ294EcoGlgC toward the substrates and the divalent ion cofactor when assayed in presence of each allosteric activating compound (Figure 6). Thus, saturation curves for UTP, Glc-1P and Mg2+ were conducted in presence of either 7.5 mM 3-PGA, 50 mM Pyr or 10 mM Fru-1,6-bisP. Figure 6a shows how the relative apparent affinity for each substrate/cofactor was affected by the respective allosteric activator. In general, the allosteric effectors decreased the S0.5 values (increasing the apparent affinity) for substrates of the chimeric enzyme. Notably, Pyr and Fru-1,6-bisP doubled the apparent affinity of the hybrid protein for UTP (the latter with an increment in the sigmoidal behavior). Pyr and 3-PGA also doubled the relative affinity for Glc-1P. On the other hand, the affinity of the chimeric enzyme for Mg2+ was not affected by 3-PGA, but it was augmented two- to three-fold by Pyr or Fru-1,6-bis P (Figure 6a). Also, the Vmax of the chimeric enzyme in the absence of an allosteric effector (15.4 U/mg, see Table 1) was enhanced to 24.6, 37.0 or 40.1 U/mg by Fru-1,6-bisP, Pyr or 3-PGA, respectively. The combination of the increase in affinity and of Vmax exerted by the allosteric effectors on the chimeric enzyme determines that they enhance the ratio Vmax/S0.5, which measures catalytic efficiency (Figure 6b), between two- and five-fold. These results highlight the sensitivity to allosteric effectors acquired in the chimeric SmuGalU-Δ294EcoGlgC.
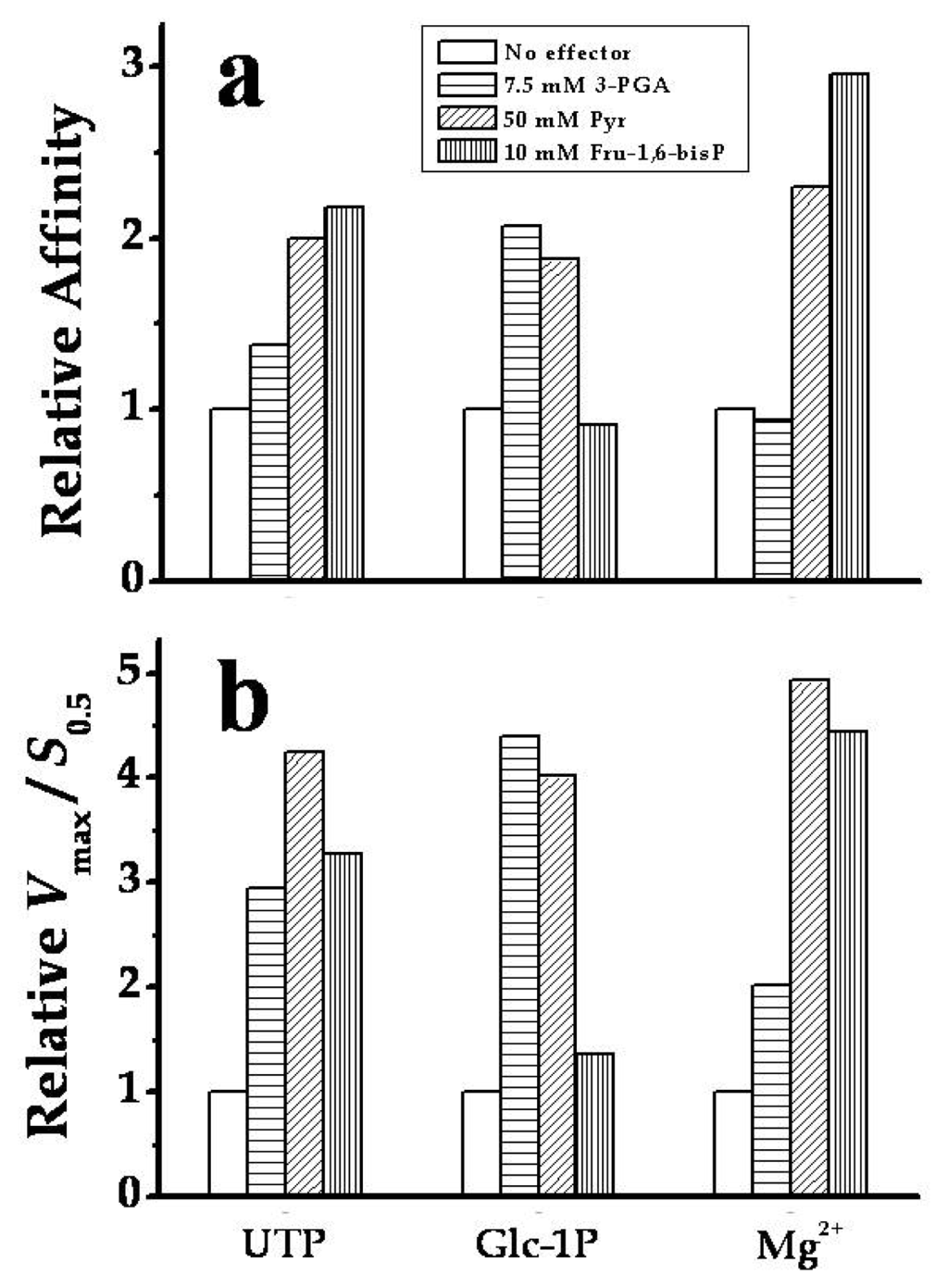
Figure 6.
Modification by allosteric activators of SmuGalU-Δ294EcoGlgC (a) substrates apparent affinities; (b) catalytic efficiencies (Vmax/S0.5). The enzyme was analyzed with no effector (white bars); 7.5 mM 3-PGA (horizontal line bar), 50 mM Pyr (oblique line bar) or 10 mM Fru-1,6-bisP (vertical line bar). Relative affinity in (a) measures the ratio between the S0.5 values determined in the absence over that in the presence of the stated amount of effector.
On the basis of the amino acid sequence alignment shown in Figure S2, we constructed a homology model for the chimeric SmuGalU-Δ294EcoGlgC protein (Figure 7). The structure was modeled using four simultaneous templates. Two templates representing the known atomic coordinates of prokaryotic (C. glutamicum and H. pylori) UDP-Glc PPases modeled the SmuGalU domain from the N-terminus of the chimeric protein. The C. glutamicum UDP-Glc PPase structure particularly allowed the location of the product UDP-Glc in the model. Then again, the architecture of the C-terminal domain of the hybrid protein (Δ294EcoGlgC) was fitted from the templates corresponding to known structures determined for two ADP-Glc PPases (the enzyme from A. tumefaciens and the small subunit from the potato tuber enzyme). The A. tumefaciens structure was mostly good for this, since it is a prokaryotic enzyme like E. coli GlgC, whereas the potato tuber structure was useful to locate the allosteric ligand in the chimeric protein. Interestingly, the model depicted in Figure 7 shows consistency with the biochemical properties determined for the chimeric protein. Thus, the predicted structure arranges a correct folding for an UDP-Glc PPase catalytic domain (including a UDP-Glc binding site) as well as a functional spatial distribution of the C-term allosteric domain, resembling its allocation in ADP-Glc PPases (Figure 7). When the C-domain is added to the N-terminal domain, it creates a pocket that could serve to accommodate binding to small molecule regulators. This is illustrated by the modeling of two sulfates that in the potato tuber enzyme are located in the inhibitory phosphate site interacting with residues in the C-terminus that are responsible for activation [].
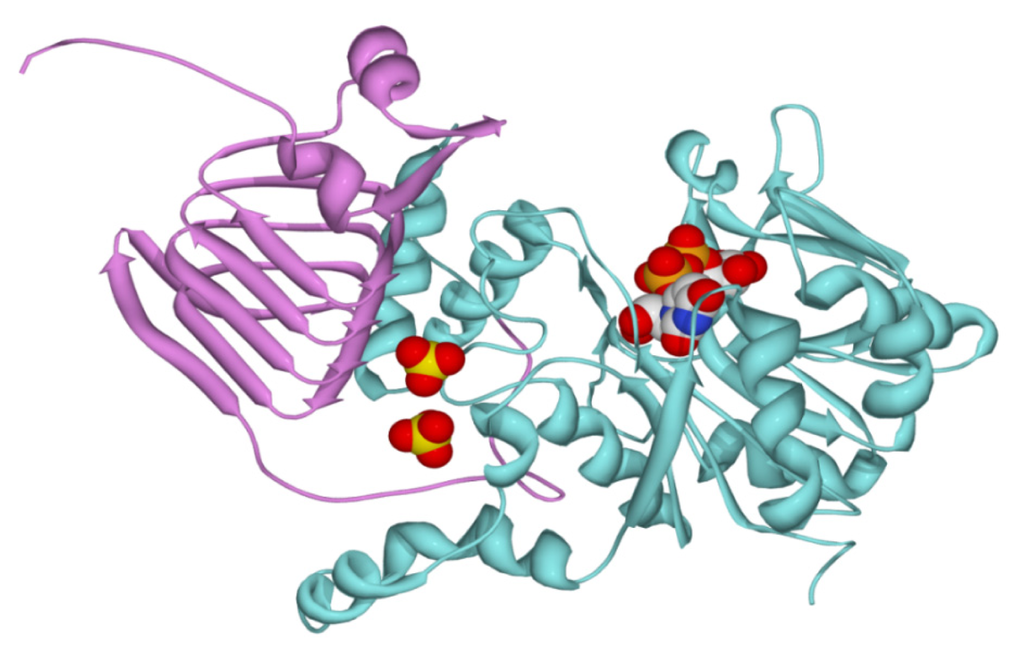
Figure 7.
Modeling of the chimeric enzyme. The model was obtained as indicated in the Experimental Section. The UDP-Glc PPase domain from S. mutans is depicted in cyan, whereas the C-domain from E. coli ADP-Glc PPase is depicted in pink. Two sulfates modeled from the structure of the potato tuber ADP-Glc PPase are shown, together with the product UDP-Glc.
3. Experimental Section
3.1. Chemicals
Antibiotics, isopropyl-β-thiogalactoside (IPTG), oligonucleotides, UTP, Glc-1P, 3-PGA, Fru-1,6-bisP and Pyr were obtained from Sigma-Aldrich (St. Louis, MO, USA). All other chemicals were of the highest quality available.
3.2. Bacteria and Plasmids
E. coli Top 10 F’ cells and pGEM®T Easy vector were used for cloning purposes. Expression of galU and qgalU was performed using pRSET-B vector (Invitrogen, Carlsbad, CA, USA) and E. coli BL21 (DE3) as host. In addition, galU was also expressed using pET24 vector (Novagen, Madison, WI, USA). DNA manipulations and E. coli cultures as well as transformations were performed according to standard protocols [].
3.3. Amplification of galU Gene from S. mutans and Construction of Chimeric galU
The 921 pb gene coding for UDP-GlcPPase, SmugalU, was amplified using S. mutans ATCC 25175 genomic DNA as a template and gene specific Smu1/Smu2 primers, designed according to available information in the GenBank database [] for S. mutans UA159 for gene coding UDP-Glc PPase (Gene ID: 1029376). All oligonucleotides used are detailed in Table 2. The 1350 pb gene coding for the chimeric protein was obtained by overlap extension PCR []. In a first step, two independent PCR reactions were conducted, using the pair of primers Smu1/Qmr2 (reaction 1) and the pair Qmr1/Qmr3 (Reaction 2). Both Qmr1 and Qmr2 primers were designed in order to hybridize SmugalU and EcoglgC region, respectively, starting at the codon for Pro295, with extra nucleotides to allow fragment fusion. Reaction 1 was conducted using a plasmid harboring SmugalU, while Reaction 2 was used as a template for the plasmid containing the glgC gene from E. coli. Products from Reaction 1 and Reaction 2 were purified and then were used together as templates in a final PCR step, using Smu1/Qmr3 primers to obtain the complete chimeric gene.

Table 2.
Oligonucleotide primers employed to amplify SmugalU gene and to construct the gene coding for the SmuGalU-Δ294EcoGlgC chimeric protein. Restriction sites are underlined.
All PCR reaction mixtures (50 μL) contained 100 ng of genomic DNA, 2 pg of each primer; 0.2 mM of each dNTP; 1.5 mM Mg2+ and 1U Taq or Pfu DNA polymerase (Fermentas, St. Leon-Rot, Germany). Standard conditions of PCR were used for 30 cycles: denaturation at 94 °C for 1 min; annealing at 55 °C for 1 min and extension at 72 °C for 2 or 4 min (depending if Taq or Pfu were used, respectively) with a final extension of 10 min at 72 °C. PCR reaction mixtures were electrophoretically defined in a 1% (w/v) agarose gel and purified with Wizard SV gel & PCR Clean Up system (Promega, Fitchburg, WI, USA). according to the manufacter’s instructions.
3.4. Cloning of SmugalU and Chimeric SmugalU-Δ294EcoglgC Genes
Amplified genes were cloned into the T-tailed plasmid pGEM-TEasy and identities were confirmed by DNA sequencing. The SmugalU gene was sub-cloned into pRSET-B BamHI/EcoRI sites to achieve a His-tagged protein at N-terminal. The chimeric gene SmugalU-Δ294EcoglgC was inserted between BamHI/XhoI from pRSET-B. Thus, both SmuGalU and the SmuGalU-Δ294EcoGlgC enzyme could be obtained with an N-terminal His-tag (see Figure 1c).
3.5. Enzymes Expressions and Purifications
Plasmid harboring SmugalU was used to transform E. coli BL21 (DE3) competent cells. Transformed cells were grown in YT2X medium at 37 °C, 200 rpm, until the A600 nm reached 0.6–0.8 and were induced with 0.8 mM IPTG at 25 °C overnight. Instead, soluble expression of chimeric SmuGalU-Δ294EcoGlgC was achieved using E. coli BL21 (DE3) pLysS as a host strain and inducing with 0.4 mM IPTG at 25 °C overnight in LB medium.
For expressing purposes, 1 L cultures in conditions detailed above were grown for each protein. Cells were harvested by centrifugation at 5000 rpm for 10 min at 4 °C and resuspended in 5 mL of buffer H (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM imidazole) per g of cells. Supernatants were obtained after cell disruption by sonication on ice, eight times for 30 s with 60 s intervals and centrifugation at 16,000 rpm for 20 min at 4 °C.
SmuGalU and chimeric SmuGalU-Δ294EcoGlgC were expressed as N-terminus His-tag fusions, in order to facilitate their subsequent purification. Enzymes were purified by affinity chromatography, using Ni-NTA Agarose resin (Invitrogen, Carlsbad, CA, USA) according to the protocol supplied by the manufacturer. Briefly, crude extract fractions prepared in buffer H were loaded onto previously equilibrated columns. After extensively washing with buffer H, samples were eluted by means of a lineal gradient to buffer I (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 300 mM imidazole). Elution fractions containing the corresponding recombinant enzyme were analyzed by SDS-PAGE [] to check for purity. For each recombinant enzyme, active fractions eluted from the Ni-NTA column were pooled, dialyzed to remove imidazole and supplemented with 10% (v/v) glycerol. Both recombinant enzymes were stable for at least six months when stored at −80 °C under the above specified respective conditions.
3.6. Protein Methods
Protein concentration was determined by the modified Bradford assay [] using bovine serum albumin as a standard. Recombinant proteins and purification fractions were defined electrophoreticaly in sodium dodecyl sulphate polyacrylamide gels (SDS-PAGE) according to []. Gels were stained with CoomassieBrilliant Blue.
3.7. Determination of Activity Optimal pH
Bis-Tris-propane [2,2′-(Propane-1,3-diyldiimino)bis[2-(hydroxymethyl)propane-1,3-diol] (Sigma, St. Louis, MO, USA), which has a wide buffering range (from pH 6.0 to pH 10.0), and tricine (Sigma, St. Louis, MO, USA) N-(2-Hydroxy-1,1-bis(hydroxymethyl)ethyl)glycine (from pH 7.5 to pH 9.0) were used to calculate the optimal pH for SmuGalU and chimeric SmuGalU-Δ294EcoGlgC activities. Measures were conducted in the UDP-Glc synthesis way.
3.8. Molecular Mass Determination
The molecular mass of the SmuGalU and the chimeric SmuGalU-Δ294EcoGlgC enzyme were determined by gel filtration using a Tricorn 5/200 column (GE Healthcare). A Gel Filtration Calibration Kit-High Molecular Weight (GE Healthcare) with protein standards including thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa) and ovoalbumin (44 kDa) was used. The column void volume was determined using a dextran blue loading solution (Promega, Fitchburg, WI, USA).
3.9. Enzyme Assays
Synthesis of UDP-Glc was assayed by following the formation of Pi (after hydrolysis of PPi by inorganic pyrophosphatase) by the highly sensitive colorimetric method previously described []. The standard reaction mixture contained 100 mM MOPS (pH 8.0), 3 mM MgCl2, 1.5 mM UTP, 0.2 mg/mL bovine serum albumin, 0.5 mU/μL yeast inorganic pyrophosphatase and a proper enzyme dilution. Assays were initiated by the addition of Glc-1P in a total volume of 50 μL. Reaction mixtures were incubated for 10 min at 37 °C and terminated by adding the Malachite Green reactive []. The complex formed with the released Pi was measured at 630 nm with an ELISA EMax detector (Molecular Devices) and using (sodium PPi) as standard.
One unit (U) of enzyme activity is equal to 1 μmol of product formed per minute under the respective assay conditions specified above.
3.10. Calculation of Kinetic Constants
Kinetic assays were performed using specified concentrations and conditions for all reaction mixture components. Saturation curves were performed by assaying the respective enzyme activity at saturating level of a fixed substrate and different concentrations of the variable substrate. The experimental data were plotted as enzyme activity (U/mg) versus substrate (or effector) concentration (mM), and kinetic constants were determined by fitting the data to the Hill equation as described elsewhere []. Fitting was performed with the Levenberg-Marquardt nonlinear least-squares algorithm provided by the computer program Origin™. Hill plots were used to calculate the Hill coefficient (nH), the maximal velocity (Vmax), and the kinetic constants that correspond to the activator, substrate or inhibitor concentrations giving 50% of the maximal activation (A0.5), velocity (S0.5) or inhibition (I0.5). All kinetic constants are the mean of at least three sets of data, which were reproducible within ±10%.
3.11. Homology Modeling
Modeling of the chimeric enzyme was performed using the program Modeller 9v1 []. For that purpose, the structure was modeled using four simultaneous templates; the known atomic coordinates of the UDP-Glc PPase from C. glutamicum in complex with magnesium and UDP-Glc (Chain A, PDB code 2pa4), the apo structure of the UDP-Glc PPase from H. pylori (chain A, PDB code 3juj), the structure of the ADP-Glc PPase from A. tumefaciens (PDB code 3brk), and the structure of potato tuber ADP-Glc PPase in complex sulfates in the putative regulatory site. In this way, the coordinates of the UDP-Glc PPases provide the structural information to build the S. mutans domain of the chimeric enzyme (N-terminus), and the ADP-Glc PPases the information needed to build the E. coli domain of the chimeric (C-terminus). The A. tumefaciens structure is particularly good for this, since it is a bacterial enzyme like E. coli, but the potato tuber structure provides information about where the sulfate ligands will go. Similarly, the C. glutamicum was used to place the product UDP-Glc in the model. The reliability of the model was evaluated using the programs Verify3D []. All the templates were manually structurally aligned to each other before the sequence alignment was performed with the chimeric target. The validation of the model with Verify3D was already good and further iterations of the alignment were not necessary.
4. Conclusions
Herein, we present results that strongly support that fusion of the C-terminal domain of E. coli ADP-Glc PPase to the entire UDP-Glc PPase from S. mutans produces a chimeric protein with UDP-Glc PPase activity sensitive to allosteric activation by specific metabolites. To the best of our knowledge this is the first report on allostery by binding of small molecules to a UDP-Glc PPase (even more in an NDP-Glc PPase distinct of ADP-Glc PPase) from prokaryotic or eukaryotic origin []. Previous work has demonstrated that UDP-Glc PPases are poorly or not regulated, with exceptions of: (i) The barley enzyme, the activity of which is affected by the occurrence of different quaternary arrangements, being active as a monomer [–]; and (ii) the E. histolytica enzyme that responds to regulation after modification by oxidants and reducing agents of critical cysteine residues [].
ADP-Glc PPases are allosteric enzymes catalyzing the key step in glycogen and starch synthesis in bacteria and plants, respectively [,]. Many studies have been conducted in order to identify the amino acidic residues involved in the allosteric response. Thus, it has been proposed that Lys39 is important in the interaction with Fru-1,6-bisP with E. coli ADP-Glc PPase [–]. In addition, Arg32 from A. tumefaciens ADP-Glc PPase (similar to Lys39 in the E. coli enzyme), Arg33, and Arg45 are involved in allosteric effector binding []. In this case, it can be seen that these residues belong to the N-terminal domain of the bacterial ADP-Glc PPase, and a recent study has clearly established the importance of the E. coli ADP-Glc PPase N-terminal domain in the activation of the enzyme []. In addition, results where the first amino acidic residues from the N-terminal domains were also removed showed the enzyme to be fully active without the allosteric activator [,] However, results obtained with chimeric enzymes obtained after switching the N- and C-terminal domains from E. coli and A. tumefaciens ADP-Glc PPases indicated that the C-terminal region was critical in determining the affinity and specificity to effectors [,]. This also suggests that in prokaryotic ADP-Glc PPases both N- and C-terminal domains are interconnected in the response of the enzyme to the allosteric effectors. In this context, our results with the chimeric enzyme SmuGalU-Δ294EcoGlgC support this model, since we clearly demonstrated that the presence of a single C-terminal domain causes sensitivity to an allosteric activator to a previously non-allosteric UDP-Glc PPase.
It has to be remarked that the activation pattern exhibited by chimeric SmuGalU-Δ294EcoGlgC is a novel feature acquired by the S. mutans UDP-Glc PPase after being transformed (by domain fusion) in the hybrid enzyme. The latter strongly supports the functional allosteric regulatory role of the C-terminal domain found in ADP-Glc PPase. Results also support the view that such a domain can be modularly fused to add allosteric properties to different PPases. It is tempting to speculate that evolution followed a similar strategy to modify one enzymes of such type (with specificity toward ADP-Glc), elongating the protein to trigger allosteric regulation in a general way. Later, critical changes in the C regulatory domain would confer specificity for different allosteric effectors within ADP-Glc PPases. We do not know the mechanism by which the N-terminal domain is activated in presence of the C-terminal domain and the proper activator, but our experiments indicate that the catalytic domain of NDP-Glc PPases may have an intrinsic property to have their activity modulated in presence of certain interactions, even when they are not natively regulated. In this case, the presence of a foreign domain triggers the allosteric properties.
Supplementary Information
ijms-14-09703-s001.pdfAcknowledgments
This work was supported by grants to AAI from CONICET (PIP 2519 and CONICET-NSF 19537/28/06/12), UNL (CAI+D Orientado and Redes), and ANPCyT (PICT’08 1754]; and to MAB from the NSF (MCB 1024945). MDAD and ACE are Fellows from CONICET. SAG and AAI are Investigators from the same Institution.
Conflict of Interest
The authors declare no conflict of interest.
References
- Ballicora, M.A.; Iglesias, A.A.; Preiss, J. ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis. Microbiol. Mol. Biol. Rev 2003, 67, 213–225. [Google Scholar]
- Ballicora, M.A.; Iglesias, A.A.; Preiss, J. ADP-glucose pyrophosphorylase: A regulatory enzyme for plant starch synthesis. Photosynth. Res 2004, 79, 1–24. [Google Scholar]
- Liu, H.W.; Thorson, J.S. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu. Rev. Microbiol 1994, 48, 223–256. [Google Scholar]
- Moretti, R.; Thorson, J.S. Enhancing the latent nucleotide triphosphate flexibility of the glucose-1-phosphate thymidylyltransferase RmlA. J. Biol. Chem 2007, 282, 16942–16947. [Google Scholar]
- Kleczkowski, L.A.; Geisler, M.; Ciereszko, I.; Johansson, H. UDP-glucose pyrophosphorylase. An old protein with new tricks. Plant Physiol 2004, 134, 912–918. [Google Scholar]
- Mollerach, M.; Garcia, E. The galU gene of Streptococcus pneumoniae that codes for a UDP-glucose pyrophosphorylase is highly polymorphic and suitable for molecular typing and phylogenetic studies. Gene 2000, 260, 77–86. [Google Scholar]
- Martinez, L.I.; Piattoni, C.V.; Garay, S.A.; Rodrigues, D.E.; Guerrero, S.A.; Iglesias, A.A. Redox regulation of UDP-glucose pyrophosphorylase from Entamoeba histolytica. Biochimie 2011, 93, 260–268. [Google Scholar]
- Asencion Diez, M.D.; Peiru, S.; Demonte, A.M.; Gramajo, H.; Iglesias, A.A. Characterization of recombinant UDP- and ADP-glucose pyrophosphorylases and glycogen synthase to elucidate glucose-1-phosphate partitioning into oligo- and polysaccharides in Streptomyces coelicolor. J. Bacteriol 2012, 194, 1485–1493. [Google Scholar]
- Mollerach, M.; Lopez, R.; Garcia, E. Characterization of the galU gene of Streptococcus pneumoniae encoding a uridine diphosphoglucose pyrophosphorylase: A gene essential for capsular polysaccharide biosynthesis. J. Exp. Med 1998, 188, 2047–2056. [Google Scholar]
- Bonofiglio, L.; Garcia, E.; Mollerach, M. Biochemical characterization of the pneumococcal glucose 1-phosphate uridylyltransferase (GalU) essential for capsule biosynthesis. Curr. Microbiol 2005, 51, 217–221. [Google Scholar]
- Bosco, M.B.; Machtey, M.; Iglesias, A.A.; Aleanzi, M. UDPglucose pyrophosphorylase from Xanthomonas spp. Characterization of the enzyme kinetics, structure and inactivation related to oligomeric dissociation. Biochimie 2009, 91, 204–213. [Google Scholar]
- Marques, A.R.; Ferreira, P.B.; Sa-Correia, I.; Fialho, A.M. Characterization of the ugpG gene encoding a UDP-glucose pyrophosphorylase from the gellan gum producer Sphingomonas paucimobilis ATCC 31461. Mol. Genet. Genomics 2003, 268, 816–824. [Google Scholar]
- Hossain, S.A.; Tanizawa, K.; Kazuta, Y.; Fukui, T. Overproduction and characterization of recombinant UDP-glucose pyrophosphorylase from Escherichia coli K-12. J. Biochem 1994, 115, 965–972. [Google Scholar]
- Thoden, J.B.; Holden, H.M. The molecular architecture of glucose-1-phosphate uridylyltransferase. Protein Sci 2007, 16, 432–440. [Google Scholar]
- Aragao, D.; Fialho, A.M.; Marques, A.R.; Mitchell, E.P.; Sa-Correia, I.; Frazao, C. The complex of Sphingomonas elodea ATCC 31461 glucose-1-phosphate uridylyltransferase with glucose-1-phosphate reveals a novel quaternary structure, unique among nucleoside diphosphate-sugar pyrophosphorylase members. J. Bacteriol 2007, 189, 4520–4528. [Google Scholar]
- Thoden, J.B.; Holden, H.M. Active site geometry of glucose-1-phosphate uridylyltransferase. Protein Sci 2007, 16, 1379–1388. [Google Scholar]
- Cupp-Vickery, J.R.; Igarashi, R.Y.; Perez, M.; Poland, M.; Meyer, C.R. Structural analysis of ADP-glucose pyrophosphorylase from the bacterium Agrobacterium tumefaciens. Biochemistry 2008, 47, 4439–4451. [Google Scholar]
- Jin, X.; Ballicora, M.A.; Preiss, J.; Geiger, J.H. Crystal structure of potato tuber ADP-glucose pyrophosphorylase. Embo. J 2005, 24, 694–704. [Google Scholar]
- Ballicora, M.A.; Sesma, J.I.; Iglesias, A.A.; Preiss, J. Characterization of chimeric ADPglucose pyrophosphorylases of Escherichia coli and Agrobacterium tumefaciens. Importance of the C-terminus on the selectivity for allosteric regulators. Biochemistry 2002, 41, 9431–9437. [Google Scholar]
- Ballicora, M.A.; Erben, E.D.; Yazaki, T.; Bertolo, A.L.; Demonte, A.M.; Schmidt, J.R.; Aleanzi, M.; Bejar, C.M.; Figueroa, C.M.; Fusari, C.M.; et al. Identification of regions critically affecting kinetics and allosteric regulation of the Escherichia coli ADP-glucose pyrophosphorylase by modeling and pentapeptide-scanning mutagenesis. J. Bacteriol 2007, 189, 5325–5333. [Google Scholar]
- Gomez-Casati, D.F.; Igarashi, R.Y.; Berger, C.N.; Brandt, M.E.; Iglesias, A.A.; Meyer, C.R. Identification of functionally important amino-terminal arginines of Agrobacterium tumefaciens ADP-glucose pyrophosphorylase by alanine scanning mutagenesis. Biochemistry 2001, 40, 10169–10178. [Google Scholar]
- Bejar, C.M.; Ballicora, M.A.; Iglesias, A.A.; Preiss, J. ADPglucose pyrophosphorylase’s N-terminus: Structural role in allosteric regulation. Biochem. Biophys. Res. Commun 2006, 343, 216–221. [Google Scholar]
- Meyer, C.R.; Bork, J.A.; Nadler, S.; Yirsa, J.; Preiss, J. Site-directed mutagenesis of a regulatory site of Escherichia coli ADP-glucose pyrophosphorylase: The role of residue 336 in allosteric behavior. Arch. Biochem. Biophys 1998, 353, 152–159. [Google Scholar]
- Meyer, C.R.; Yirsa, J.; Gott, B.; Preiss, J. A kinetic study of site-directed mutants of Escherichia coli ADP-glucose pyrophosphorylase: The role of residue 295 in allosteric regulation. Arch. Biochem. Biophys 1998, 352, 247–254. [Google Scholar]
- Bejar, C.M.; Ballicora, M.A.; Gomez-Casati, D.F.; Iglesias, A.A.; Preiss, J. The ADP-glucose pyrophosphorylase from Escherichia coli comprises two tightly bound distinct domains. FEBS Lett 2004, 573, 99–104. [Google Scholar]
- Ajdic, D.; McShan, W.M.; McLaughlin, R.E.; Savic, G.; Chang, J.; Carson, M.B.; Primeaux, C.; Tian, R.; Kenton, S.; Jia, H.; et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 2002, 99, 14434–14439. [Google Scholar]
- Bonofiglio, L.; Garcia, E.; Mollerach, M. The galU gene expression in Streptococcus pneumoniae. FEMS Microbiol. Lett 2012, 332, 47–53. [Google Scholar]
- Varon, D.; Boylan, S.A.; Okamoto, K.; Price, C.W. Bacillus subtilis gtaB encodes UDP-glucose pyrophosphorylase and is controlled by stationary-phase transcription factor sigma B. J. Bacteriol 1993, 175, 3964–3971. [Google Scholar]
- Kim, H.; Choi, J.; Kim, T.; Lokanath, N.K.; Ha, S.C.; Suh, S.W.; Hwang, H.Y.; Kim, K.K. Structural basis for the reaction mechanism of UDP-glucose pyrophosphorylase. Mol. Cells 2010, 29, 397–405. [Google Scholar]
- Iglesias, A.A.; Ballicora, M.A.; Sesma, J.I.; Preiss, J. Domain swapping between a cyanobacterial and a plant subunit ADP-glucose pyrophosphorylase. Plant Cell Physiol 2006, 47, 523–530. [Google Scholar]
- Silva, E.; Marques, A.R.; Fialho, A.M.; Granja, A.T.; Sa-Correia, I. Proteins encoded by Sphingomonas elodea ATCC 31461 rmlA and ugpG genes, involved in gellan gum biosynthesis, exhibit both dTDP- and UDP-glucose pyrophosphorylase activities. App. Environ. Microbiol 2005, 71, 4703–4712. [Google Scholar]
- Weissborn, A.C.; Liu, Q.; Rumley, M.K.; Kennedy, E.P. UTP: alpha-d-glucose-1-phosphate uridylyltransferase of Escherichia coli: Isolation and DNA sequence of the galU gene and purification of the enzyme. J. Bacteriol 1994, 176, 2611–2618. [Google Scholar]
- Asencion Diez, M.D.; Demonte, A.; Giacomelli, J.; Garay, S.; Rodrigues, D.; Hofmann, B.; Hecht, H.J.; Guerrero, S.A.; Iglesias, A.A. Functional characterization of GDP-mannose pyrophosphorylase from Leptospira interrogans serovar Copenhageni. Arch. Microbiol 2010, 192, 103–114. [Google Scholar]
- Ning, B.; Elbein, A.D. Purification and properties of mycobacterial GDP-mannose pyrophosphorylase. Arch. Biochem. Biophys 1999, 362, 339–345. [Google Scholar]
- Frueauf, J.B.; Ballicora, M.A.; Preiss, J. ADP-glucose pyrophosphorylase from potato tuber: Site-directed mutagenesis of homologous aspartic acid residues in the small and large subunits. Plant J 2003, 33, 503–511. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, New York, NY, USA, 2001. [Google Scholar]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res 2005, 33, D34–D38. [Google Scholar]
- Higuchi, R.; Krummel, B.; Saiki, R.K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res 1988, 16, 7351–7367. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [Google Scholar]
- Fusari, C.; Demonte, A.M.; Figueroa, C.M.; Aleanzi, M.; Iglesias, A.A. A colorimetric method for the assay of ADP-glucose pyrophosphorylase. Anal. Biochem 2006, 352, 145–147. [Google Scholar]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol 1993, 234, 779–815. [Google Scholar]
- Luthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar]
- Decker, D.; Meng, M.; Gornicka, A.; Hofer, A.; Wilczynska, M.; Kleczkowski, L.A. Substrate kinetics and substrate effects on the quaternary structure of barley UDP-glucose pyrophosphorylase. Phytochemistry 2012, 79, 39–45. [Google Scholar]
- Kleczkowski, L.A.; Martz, F.; Wilczynska, M. Factors affecting oligomerization status of UDP-glucose pyrophosphorylase. Phytochemistry 2005, 66, 2815–2821. [Google Scholar]
- Martz, F.; Wilczynska, M.; Kleczkowski, L.A. Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose pyrophosphorylase. Biochem. J 2002, 367, 295–300. [Google Scholar]
- Gardiol, A.; Preiss, J. Escherichia coli E-39 ADPglucose synthetase has different activation kinetics from the wild-type allosteric enzyme. Arch. Biochem. Biophys 1990, 280, 175–180. [Google Scholar]
- Parsons, T.F.; Preiss, J. Biosynthesis of bacterial glycogen. Isolation and characterization of the pyridoxal-P allosteric activator site and the ADP-glucose-protected pyridoxal-P binding site of Escherichia coli B ADP-glucose synthase. J. Biol. Chem 1978, 253, 7638–7645. [Google Scholar]
- Parsons, T.F.; Preiss, J. Biosynthesis of bacterial glycogen. Incorporation of pyridoxal phosphate into the allosteric activator site and an ADP-glucose-protected pyridoxal phosphate binding site of Escherichia coli B ADP-glucose synthase. J. Biol. Chem 1978, 253, 6197–6202. [Google Scholar]
- Wu, M.X.; Preiss, J. The N-terminal region is important for the allosteric activation and inhibition of the Escherichia coli ADP-glucose pyrophosphorylase. Arch. Biochem. Biophys 1998, 358, 182–188. [Google Scholar]
- Wu, M.X.; Preiss, J. Truncated forms of the recombinant Escherichia coli ADP-glucose pyrophosphorylase: The importance of the N-terminal region for allosteric activation and inhibition. Arch. Biochem. Biophys 2001, 389, 159–165. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).