Single Nucleotide Polymorphisms in the Leptin-a Gene and Associations with Growth Traits in the Orange-Spotted Grouper (Epinephelus coioides)
Abstract
:1. Introduction
2. Results and Discussion
2.1. SNP Identification and Genotyping
2.2. Association Analysis with Growth Traits
2.3. Discussion
3. Experimental Section
3.1. Materials and Phenotypic Data Collection
3.2. PCR Amplification and SNP Identification
3.3. SNP Identification and Genotyping
3.4. Statistical Analysis
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Nelson, J.S. Fishes of the World; John Wiley and Sons: New York, NY, USA; p. 1994.
- Heemstra, P.C.; Randall, J.E. FAO Fisheries Synopsis: Groupers of the World (Family Serranidae, Subfamily Epinephelinae); Food and Agriculture Organization of the United Nations: Rome, Italy, 1993; Volume 16, pp. 1–382. [Google Scholar]
- Tupper, M.; Sheriff, N. Capture-based Aquaculture of Groupers. In FAO Fisheries Technical Paper; FAO: Rome, Italy, 2008; pp. 217–253. [Google Scholar]
- FAO. Cultured Aquatic Species Information Programme, Epinephelus coioides. Available online: http://www.fao.org (accessed on 28 June 2011).
- Zhang, Y.Y.; Proenca, R.; Maffei, M.; Barone, M.; Leopold, L.; Friedman, J.M. Positional cloning of the mouse obese gene and its human homologue. Nature 1994. [Google Scholar] [CrossRef]
- Houseknecht, K.L.; Baile, C.A.; Matteri, R.L.; Spurlock, M.E. The biology of leptin: A review. J. Anim. Sci 1998, 76, 1405–1420. [Google Scholar]
- Mácajová, M.; Lamosova, D.; Zeman, M. Role of leptin in farm animals: A review. J. Vet. Med. A Physiol. Pathol. Clin. Med 2004, 51, 157–166. [Google Scholar]
- Moschos, S.; Chan, J.L.; Mantzoros, C.S. Leptin and reproduction: A review. Fertil. Steril 2002, 77, 433–444. [Google Scholar]
- Kurokawa, T.; Uji, S.; Suzuki, T. Identification of cDNA coding for a homologue to mammalian leptin from pufferfish, Takifugu rubripes. Peptides 2005, 26, 745–750. [Google Scholar]
- Huising, M.O.; Geven, E.J.; Kruiswijk, C.P.; Nabuurs, S.B.; Stolte, E.H.; Spanings, F.A.; Verburg-van, K.B.; Flik, G. Increased leptin expression in common Carp (Cyprinus carpio) after food intake but not after fasting or feeding to satiation. Endocrinology 2006, 147, 5786–5797. [Google Scholar]
- Gorissen, M.; Bernier, N.J.; Nabuurs, S.B.; Flik, G.; Huising, M.O. Two divergent leptin paralogues in zebrafish (Danio rerio) that originate early in teleostean evolution. J. Endocrinol 2009, 201, 329–339. [Google Scholar]
- Kurokawa, T.; Murashita, K. Genomic characterization of multiple leptin genes and a leptin receptor gene in the Japanese medaka, Oryzias latipes. Gen. Comp. Endocrinol 2009, 161, 229–237. [Google Scholar]
- Li, G.; Liang, X.; Xie, Q.; Li, G.; Yu, Y.; Lai, K. Gene structure, recombinant expression and functional characterization of grass carp leptin. Gen. Comp. Endocrinol 2010, 166, 117–127. [Google Scholar]
- Murashita, K.; Uji, S.; Yamamoto, T.; Ronnestad, I.; Kurokawa, T. Production of recombinant leptin and its effects on food intake in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. B 2008, 150, 377–384. [Google Scholar]
- Renestad, I.; Nilsen, T.O.; Murashita, K.; Angotzi, A.R.; Moen, A.G.; Stefansson, S.O.; Kling, P.; Bjornsson, B.T.; Kurokawa, T. Leptin and leptin receptor genes in Atlantic salmon: Cloning, phylogeny, tissue distribution and expression correlated to long-term feeding status. Gen. Comp. Endocrinol 2010, 168, 55–70. [Google Scholar]
- Won, E.T.; Baltzegar, D.A.; Picha, M.E.; Borski, R.J. Cloning and characterization of leptin in a Perciform fish, the striped bass (Morone saxatilis): Control of feeding and regulation by nutritional state. Gen. Comp. Endocrinol 2012, 178, 98–107. [Google Scholar]
- Tinoco, A.B.; Nisembaum, L.G.; Isorna, E.; Delgado, M.J.; Pedro, N. Leptins and leptin receptor expression in the goldfish (Carassius auratus). Regulation by food intake and fasting/overfeeding conditions. Peptides 2012, 34, 329–335. [Google Scholar]
- Zhang, H.; Chen, H.P.; Zhang, Y.; Li, S.S.; Lu, D.Q.; Zhang, H.F.; Meng, Z.N.; Liu, X.C.; Lin, H.R. Molecular cloning, characterization and expression profiles of multiple leptin genes and a leptin receptor gene in orange-spotted grouper (Epinephelus coioides). Gen. Comp. Endocrinol 2013, 181, 295–305. [Google Scholar]
- Copeland, D.L.; Duff, R.J.; Liu, Q.; Prokop, J.; Londraville, R.L. Leptin in teleost fishes: An argument for comparative study. Front. Physiol 2011, 2, 1–11. [Google Scholar]
- Uemoto, Y.; Kikuchi, T.; Nakano, H.; Sato, S.; Shibata, T.; Kadowaki, H.; Katoh, K.; Kobayashi, E.; Suzuki, K. Effects of porcine leptin receptor gene polymorphisms on backfat thickness, fat area ratios by image analysis, and serum leptin concentrations in a Duroc purebred population. Anim. Sci. J 2012, 83, 375–85. [Google Scholar]
- Da, S.R.; Ferraz, J.B.; Meirelles, F.V.; Eler, J.P.; Balieiro, J.C.; Cucco, D.C.; Mattos, E.C.; Rezende, F.M.; Silva, S.L. Association of single nucleotide polymorphisms in the bovine leptin and leptin receptor genes with growth and ultrasound carcass traits in Nellore cattle. Genet. Mol. Res 2012, 11, 3721–3728. [Google Scholar] [Green Version]
- Kuss, A.W.; Gogol, J.; Geidermann, H. Associations of a polymorphic AP-2 binding site in the 5′-flanking region of the bovine beta-lactoglobulin gene with milk proteins. J. Dairy. Sci 2003, 86, 2213–2218. [Google Scholar]
- Buchanan, F.C.; Fitzsimmons, C.J.; van Kessel, A.G.; Thue, T.D.; Winkelman-Sim, D.C.; Schmutz, S.M. Association of a missense mutation in the bovine leptin gene with carcass fat content and leptin mRNA levels. Genet. Sel. Evol. 2002, 34, 105–116. [Google Scholar]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Sci. Signal 2001, 291, 1304. [Google Scholar]
- Özlem, E.; Dursun, Ö. An application of information theoretical measures for DNA structure. Turk. Klinikleri J. Biostat 2011, 3, 1–7. [Google Scholar]
- Fiume, E.; Christou, P.; Gianì, S.; Breviario, D. Introns are key regulatory elements of rice tubulin expression. Planta 2004, 218, 693–703. [Google Scholar]
- Nie, Q.; Sun, B.; Zhang, D.; Luo, C.; Ishag, N.A.; Lei, M.; Yang, G.; Zhang, X. High diversity of the chicken growth hormone gene and effects on growth and carcass traits. J. Hered 2005, 96, 698–703. [Google Scholar]
- Sun, Y.; Yu, X.; Tong, J. Polymorphisms in myostatin gene and associations with growth traits in the common carp (Cyprinus carpio L.). Int. J. Mol. Sci 2012, 13, 14956–14961. [Google Scholar]
- Zhang, S.; Li, H.; Shi, H. Single marker and haplotype analysis of the chicken apolipoprotein B gene T123G and D9500D9-polymorphism reveals association with body growth and obesity. Poult Sci 2006, 85, 178–184. [Google Scholar]
- Stachowiak, M.; Mackowski, M.; Madeja, Z.; Szydlowski, M.; Buszka, A.; Kaczmarek, P.; Rubis, B.; Mackowiak, P.; Nowak, K.W.; Switonski, M. Polymorphism of the porcine leptin gene promoter and analysis of its association with gene expression and fatness traits. Biochem. Genet 2007, 45, 245–253. [Google Scholar]
- Kulig, H.; Kmiec, M. Association between leptin gene polymorphisms and growth traits in Limousin cattle. Russ. J. Genet 2009, 45, 738–741. [Google Scholar]
- Gill, J.L.; Bishop, S.C.; McCorquodale, C.; Williams, J.L.; Wiener, P. Association of selected SNP with carcass and taste panel assessed meat quality traits in a commercial population of Aberdeen Angus-sired beef cattle. Genet. Sel. Evol 2009, 41, 36. [Google Scholar]
- Clempson, A.M.; Pollott, G.E.; Brickell, J.S.; Bourne, N.E.; Munce, N.; Wathes, D.C. Evidence that leptin genotype is associated with fertility, growth, and milk production in Holstein cows. J. Dairy. Sci 2011, 94, 3618–3628. [Google Scholar]
- Melucci, L.M.; Panarace, M.; Feula, P.; Villarreal, E.L.; Grigioni, G.; Carduza, F.; Soria, L.A.; Mezzadra, C.A.; Arceo, M.E.; Papaleo Mazzucco, J.; et al. Genetic and management factors affecting beef quality in grazing Hereford steers. Meat Sci 2012, 92, 768–774. [Google Scholar]
- Ovilo, C.; Fernández, A.; Noguera, J.L.; Barragán, C.; Letón, R.; Rodríguez, C.; Mercadé, A.; Alves, E.; Folch, J.M.; Varona, L.; Toro, M. Fine mapping of porcine chromosome 6 QTL and LEPR effects on body composition in multiple generations of an Iberian by Landrace intercross. Genet. Res 2005, 85, 57–67. [Google Scholar]
- Lei, M.; Luo, C.; Peng, X.; Fang, M.; Nie, Q.; Zhang, D.; Yang, G.; Zhang, X. Polymorphism of growth-correlated genes associated with fatness and muscle fiber traits in chickens. Poult. Sci 2007, 86, 835–842. [Google Scholar]
- Guo, Y.; Chen, H.; Lan, X.; Zhang, B.; Pan, C.; Zhang, L.; Zhang, C.; Zhao, M. Novel SNPs of the bovine LEPR gene and their association with growth traits. Biochem. Genet 2008, 46, 828–834. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 2001. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997, 25, 4876–4882. [Google Scholar]


| SNP | Position | Mutation type | Sample size | Genotype frequencies (%) | Allele frequencies (%) | Ho | He | p-value (χ2, HWE) | PIC a | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.182T > G | Intron1 | Untranslated | 200 | TT | TG | GG | T | G | 0.055 | 0.053 | 0.923 | 0.052 |
| 94.5 | 5.5 | 0 | 97.25 | 2.75 | ||||||||
| c.183G > T | Intron1 | Untranslated | 200 | GG | GT | TT | G | T | 0.055 | 0.053 | 0.923 | 0.052 |
| 94.5 | 5.5 | 0 | 97.25 | 2.75 | ||||||||
| c.339C > G | Exon2 | Synonymous | 200 | CC | CG | GG | C | G | 0.030 | 0.030 | 0.977 | 0.029 |
| T:ACC→ACG | 97.0 | 3.0 | 0 | 98.50 | 1.50 | |||||||
| c.345C > T | Exon2 | Synonymous | 200 | CC | CT | TT | C | T | 0.075 | 0.072 | 0.859 | 0.070 |
| N:AAC→AAT | 92.5 | 7.5 | 0 | 96.25 | 3.75 | |||||||
| c.447G > A | Exon2 | Synonymous | 200 | GG | GA | AA | G | A | 0.040 | 0.039 | 0.959 | 0.038 |
| P:CCG→CCA | 96.0 | 4.0 | 0 | 98.00 | 2.00 | |||||||
| c.531C > T | Exon2 | Synonymous | 200 | CC | CT | TT | C | T | 0.205 | 0.251 | 0.033* | 0.220 |
| L:CTC→CTT | 75.0 | 20.5 | 4.5 | 85.25 | 14.75 | |||||||
| SNP | Genotypes | N | BWT (g) | BWH (cm) | OL (cm) | TW (cm) | HL (cm) | CPL (cm) | K (%) |
|---|---|---|---|---|---|---|---|---|---|
| c.182 + 183 | AA(TT/GG) | 189 | 89.67 ± 3.12 a | 4.65 ± 0.05 a | 17.89 ± 0.20 | 2.73 ± 0.04 | 5.79 ± 0.07 | 2.44 ± 0.04 | 2.67 ± 0.32 |
| TG > GT | AB(TG/GT) | 11 | 121.82 ± 12.94 b | 5.17 ± 0.22 b | 19.32 ± 0.82 | 3.03 ± 0.16 | 6.23 ± 0.28 | 2.66 ± 0.15 | 2.74 ± 0.38 |
| p-value | 0.017* | 0.020* | 0.091 | 0.074 | 0.130 | 0.160 | 0.518 | ||
| c.339C > G | CC | 194 | 91.26 ± 3.12 | 4.68 ± 0.05 | 17.97 ± 0.20 | 2.75 ± 0.04 | 5.82 ± 0.07 | 2.46 ± 0.04 | 2.67 ± 0.32 |
| CG | 6 | 97.33 ± 17.77 | 4.83 ± 0.30 | 18.02 ± 1.12 | 2.83 ± 0.22 | 5.83 ± 0.38 | 2.45 ± 0.21 | 2.81 ± 0.34 | |
| p-value | 0.737 | 0.602 | 0.964 | 0.692 | 0.963 | 0.980 | 0.285 | ||
| c.345C > T | CC | 185 | 90.51 ± 3.19 | 4.67 ± 0.05 | 17.90 ± 0.20 | 2.74 ± 0.04 | 5.80 ± 0.07 | 2.45 ± 0.04 | 2.69 ± 0.32 a |
| CT | 15 | 102.93 ± 11.21 | 4.87 ± 0.19 | 18.77 ± 0.70 | 2.83 ± 0.14 | 6.03 ± 0.24 | 2.49 ± 0.13 | 2.51 ± 0.27 b | |
| p-value | 0.288 | 0.303 | 0.234 | 0.520 | 0.346 | 0.802 | 0.036* | ||
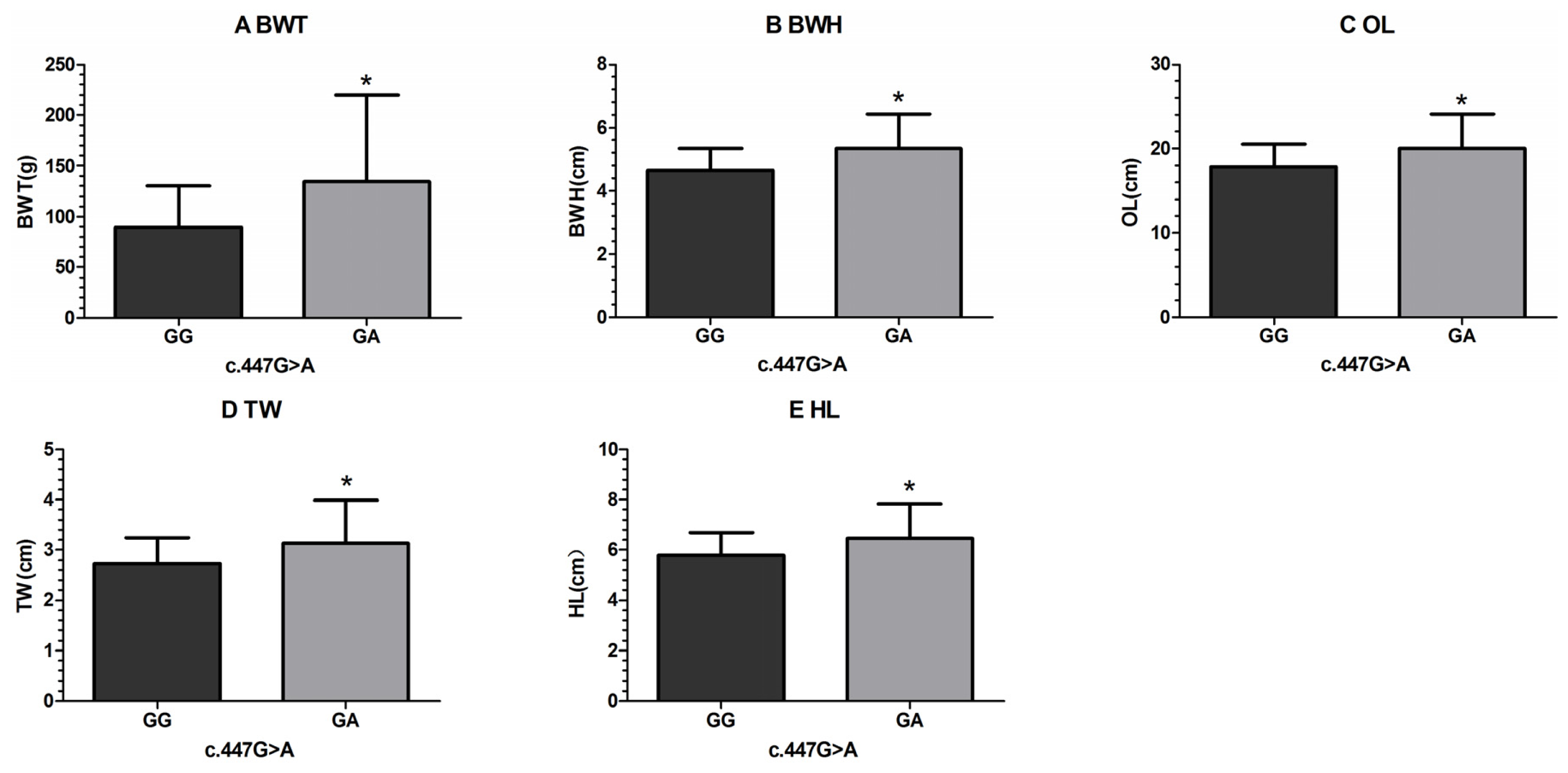
| c.447G > A | GG | 192 | 89.64 ± 3.08 a | 4.65 ± 0.05 a | 17.88 ± 0.20 a | 2.73± 0.04 a | 5.79 ± 0.07 | 2.44 ± 0.04 | 2.67 ± 0.02 |
| GA | 8 | 134.75± 15.07 b | 5.34 ± 0.25 b | 20.01 ± 0.95 b | 3.15± 0.19 b | 6.48 ± 0.33 | 2.75 ± 0.18 | 2.72 ± 0.11 | |
| p-value | 0.004* | 0.009* | 0.030* | 0.029* | 0.040* | 0.092 | 0.681 | ||
| c.531C > T | CC | 150 | 92.59 ± 3.55 | 4.70 ± 0.06 | 18.11 ± 0.22 | 2.76 ± 0.04 | 5.83 ± 0.08 | 2.48 ± 0.04 | 2.66 ± 0.30 |
| CT | 41 | 91.66 ± 6.78 | 4.71 ± 0.11 | 17.80 ± 0.42 | 2.80 ± 0.08 | 5.88 ± 0.14 | 2.41 ± 0.08 | 2.72 ± 0.38 | |
| TT | 9 | 71.33 ± 14.47 | 4.30 ± 0.24 | 16.39 ± 0.90 | 2.38 ± 0.18 | 5.32 ± 0.31 | 2.31 ± 0.17 | 2.70 ± 0.32 | |
| p-value | 0.363 | 0.273 | 0.170 | 0.092 | 0.251 | 0.507 | 0.629 | ||
| CC/CT | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | ||
| CT/TT | 0.615 | 0.372 | 0.471 | 0.094 | 0.311 | 1.000 | 1.000 | ||
| TT/CC | 0.466 | 0.343 | 0.200 | 0.116 | 0.335 | 1.000 | 1.000 |
| Phenotypic traits a | Mean | Std. Error | Std. Deviation | CV b (%) | Minimum | Maximum | Range | p-value c (K-S test) |
|---|---|---|---|---|---|---|---|---|
| BWT (g) | 91.440 | 3.070 | 43.418 | 47.48 | 20.00 | 310.00 | 290.00 | 0.000 |
| BWH (cm) | 4.681 | 0.051 | 0.726 | 15.51 | 2.70 | 7.50 | 4.80 | 0.076 * |
| OL (cm) | 17.967 | 0.193 | 2.723 | 15.16 | 12.20 | 26.30 | 14.10 | 0.057 * |
| BL (cm) | 14.774 | 0.167 | 2.368 | 16.03 | 10.00 | 22.10 | 12.10 | 0.041 |
| TW (cm) | 2.748 | 0.038 | 0.533 | 19.40 | 1.50 | 4.90 | 3.40 | 0.026 |
| HL (cm) | 5.816 | 0.065 | 0.925 | 15.91 | 3.80 | 9.00 | 5.20 | 0.000 |
| CPW (cm) | 1.606 | 0.017 | 0.243 | 15.13 | 1.10 | 2.50 | 1.40 | 0.000 |
| CPL (cm) | 2.455 | 0.036 | 0.506 | 20.59 | 1.30 | 3.80 | 2.50 | 0.000 |
| SL (cm) | 1.186 | 0.016 | 0.230 | 19.39 | 0.70 | 2.20 | 1.50 | 0.000 |
| ED (cm) | 0.946 | 0.007 | 0.093 | 9.84 | 0.60 | 1.20 | 0.60 | 0.000 |
| ID (cm) | 0.937 | 0.011 | 0.161 | 17.19 | 0.60 | 1.50 | 0.90 | 0.000 |
| K (%) | 2.676 | 0.023 | 0.320 | 11.96 | 1.78 | 3.53 | 1.75 | 0.200 * |
| Primer code | Primer sequences (5′–3′) | Product size | TM (°C) |
|---|---|---|---|
| Leptin-a1 | F: ATGGACTACACTCTGGCCCTG | 573 bp | 51.0 |
| R: TCAGCAAGTCTCAAGATGGTCC | 51.5 | ||
| Leptin-a2 | F: GGAACTACAGAACTACTTGGAA | 698 bp | 51.0 |
| R: GTGCTGGAGGAAATGTATTC | 52.0 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wei, Y.; Huang, H.; Meng, Z.; Zhang, Y.; Luo, J.; Chen, G.; Lin, H. Single Nucleotide Polymorphisms in the Leptin-a Gene and Associations with Growth Traits in the Orange-Spotted Grouper (Epinephelus coioides). Int. J. Mol. Sci. 2013, 14, 8625-8637. https://doi.org/10.3390/ijms14048625
Wei Y, Huang H, Meng Z, Zhang Y, Luo J, Chen G, Lin H. Single Nucleotide Polymorphisms in the Leptin-a Gene and Associations with Growth Traits in the Orange-Spotted Grouper (Epinephelus coioides). International Journal of Molecular Sciences. 2013; 14(4):8625-8637. https://doi.org/10.3390/ijms14048625
Chicago/Turabian StyleWei, Yun, Hai Huang, Zining Meng, Yong Zhang, Jian Luo, Guohua Chen, and Haoran Lin. 2013. "Single Nucleotide Polymorphisms in the Leptin-a Gene and Associations with Growth Traits in the Orange-Spotted Grouper (Epinephelus coioides)" International Journal of Molecular Sciences 14, no. 4: 8625-8637. https://doi.org/10.3390/ijms14048625




