Structure, Function and Networks of Transcription Factors Involved in Abiotic Stress Responses
Abstract
:1. Introduction
2. Large Plant TF Families in Abiotic Stress Responses
2.1. bZIP TFs
2.2. AP2/ERF TFs
2.3. NAC TFs
2.4. WRKY TFs
2.5. Cys2His2 Zinc Finger (C2H2 ZF) TFs
2.6. MYB TFs
2.7. bHLH TFs
2.8. Various TFs in Abiotic Stress Responses
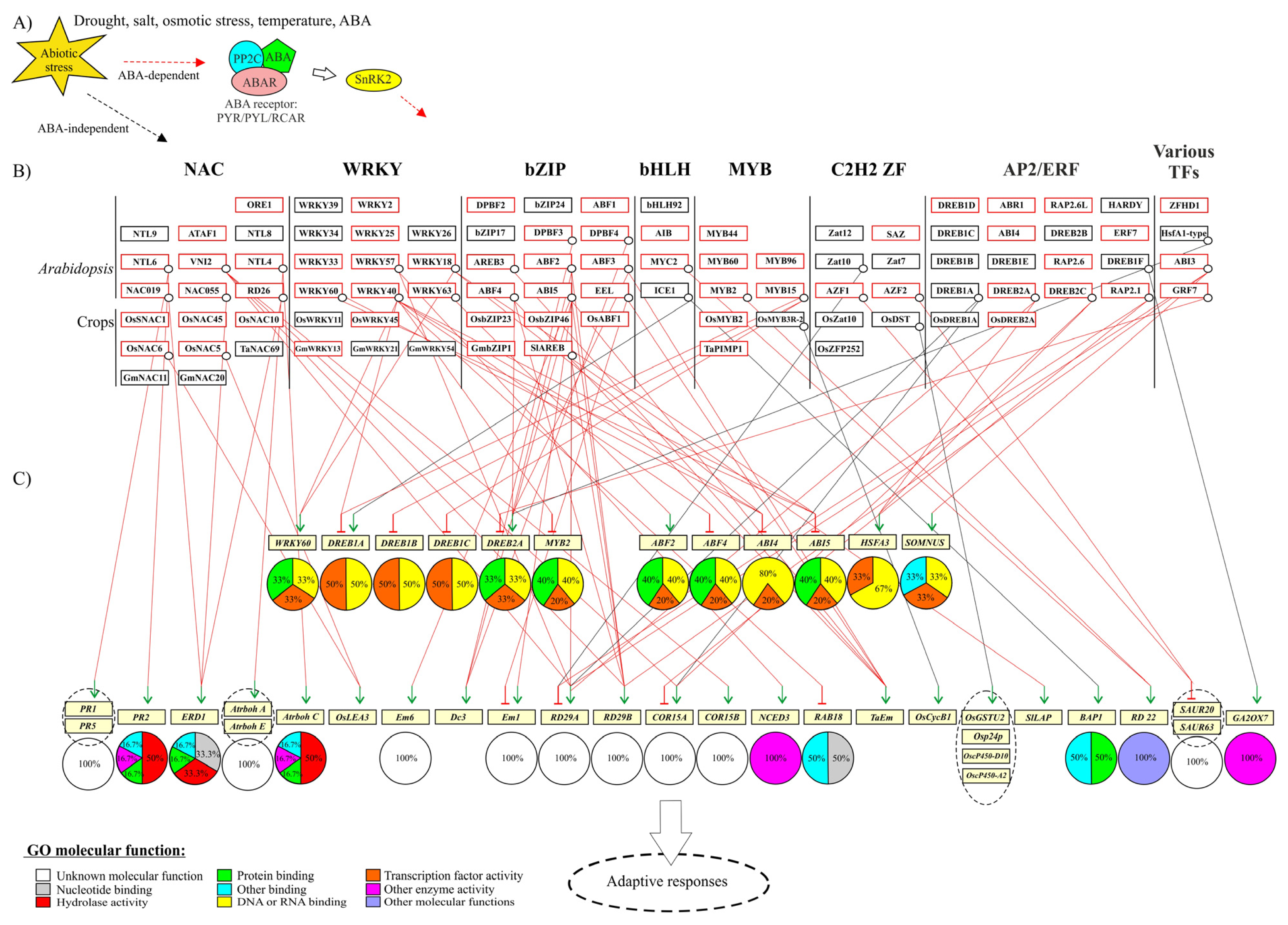
2.9. Gene Regulatory Networks in Abiotic Stress Responses
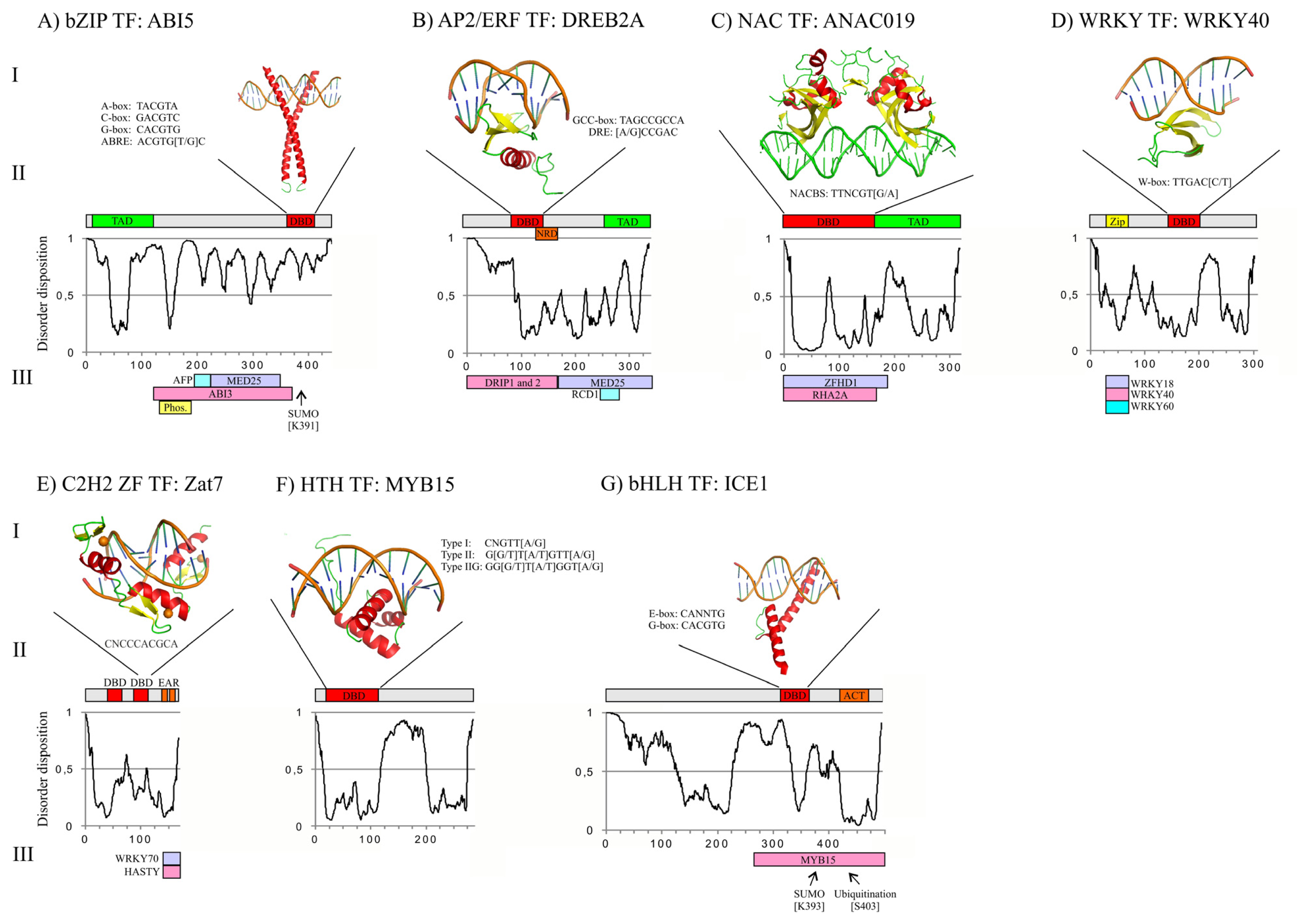
3. Structures of Abiotic Stress-Related TFs: Determinants of Function
3.1. bZIP DBD and Overall Structure of ABI5
3.2. AP2/ERF DBD and Overall Structure of DREB2A
3.3. NAC DBD and Overall Structure of ANAC019
3.4. WRKY DBD and Overall Structure of WRKY40
3.5. C2H2 ZF DBD and Overall Structure of Zat7
3.6. Helix-Turn-Helix (HTH) DBD and Overall Structure of MYB15
3.7. bHLH DBD and Overall Structure of ICE1
3.8. Structure of TFs in Abiotic Stress Responses
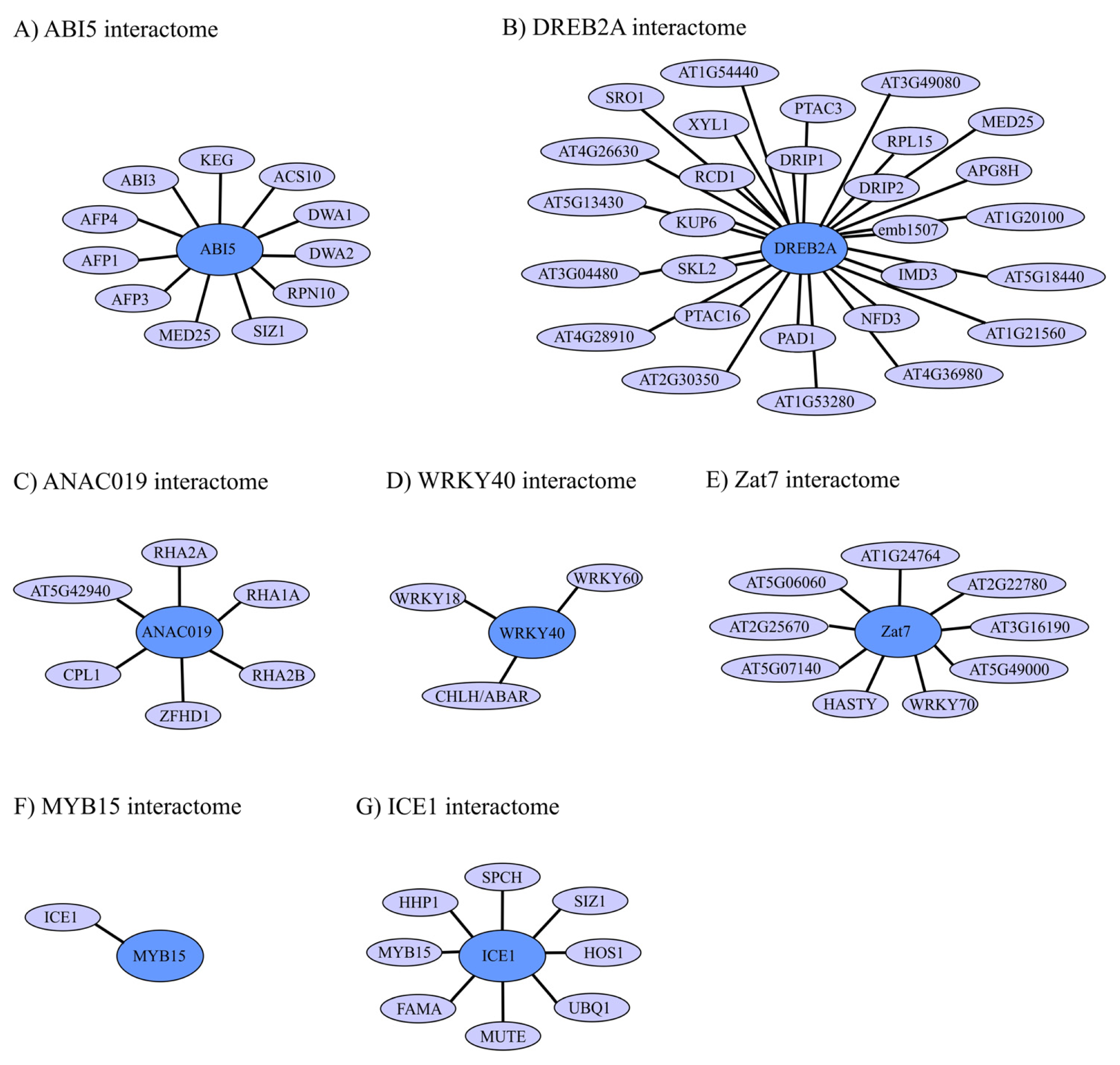
4. Regulation of TF Activity through Protein-Protein Interactions (PPIs)
4.1. ABI5 Interactome and Regulation
4.2. DREB2A Interactome and Regulation
4.3. ANAC019 Interactome and Regulation
4.4. WRKY40 Interactome and Regulation
4.5. Zat7 Interactome and Regulation
4.6. ICE1 and MYB15 Interactomes and Regulation
4.7. TF Interactomes in Abiotic Plant Stress
5. Conclusion
Acknowledgements
References
- Matsui, A.; Ishida, J.; Morosawa, T.; Mochizuki, Y.; Kaminuma, E.; Endo, T.A.; Okamoto, M.; Nambara, E.; Nakajima, M.; Kawashima, M.; et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 2008, 49, 1135–1149. [Google Scholar]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol 2010, 61, 651–679. [Google Scholar]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar]
- Udvardi, M.K.; Kakar, K.; Wandrey, M.; Montanari, O.; Murray, J.; Andriankaja, A.; Zhang, J.Y.; Benedito, V.; Hofer, J.M.; Chueng, F.; et al. Legume transcription factors: Global regulators of plant development and response to the environment. Plant Physiol 2007, 144, 538–549. [Google Scholar]
- Kilian, J.; Peschke, F.; Berendzen, K.W.; Harter, K.; Wanke, D. Prerequisites, performance and profits of transcriptional profiling the abiotic stress response. Biochim. Biophys. Acta 2012, 1819, 166–175. [Google Scholar]
- Golldack, D.; Lüking, I.; Yang, O. Plant tolerance to draught and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep 2011, 30, 1389–1391. [Google Scholar]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 2012, 1819, 120–128. [Google Scholar]
- Ciftci-Yilmaz, S.; Mittler, R. The zinc finger network of plants. Cell Mol. Life Sci 2008, 65, 1150–1160. [Google Scholar]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci 2010, 15, 573–581. [Google Scholar]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res 2011, 124, 509–525. [Google Scholar]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 97–103. [Google Scholar]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci 2012, 17, 369–381. [Google Scholar]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J 2012, 10, 2–11. [Google Scholar]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol 2010, 51, 1821–1839. [Google Scholar]
- Gene Ontology search tool. Available online: http://www.arabidopsis.org/tools/bulk/go/index.jsp accessed on 27 February 2013.
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci 2002, 7, 106–111. [Google Scholar]
- Kang, J.Y.; Choi, H.I.; Im, M.Y.; Kim, S.Y. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 2002, 14, 343–357. [Google Scholar]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 2010, 61, 672–685. [Google Scholar]
- Kim, J.S.; Mizoi, J.; Yoshida, T.; Fujita, Y.; Nakajima, J.; Ohori, T.; Todaka, D.; Nakashima, K.; Hirayama, T.; Shinozaki, K.; et al. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol 2011, 52, 2136–2146. [Google Scholar]
- Miura, K.; Lee, J.; Jin, J.B.; Yoo, C.Y.; Miura, T.; Hasegawa, P.M. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 5418–5423. [Google Scholar] [Green Version]
- Finkelstein, R.R. Maternal Effects Govern Variable Dominance of Two Abscisic Acid Response Mutations in Arabidopsis thaliana. Plant Physiol 1994, 105, 1203–1208. [Google Scholar]
- Nakashima, K.; Fujita, Y.; Katsura, K.; Maruyama, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulation of. Plant Mol Biol 2006, 60, 51–68. [Google Scholar]
- Kim, S.Y.; Ma, J.; Perret, P.; Li, Z.; Thomas, T.L. Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol 2002, 130, 688–697. [Google Scholar]
- Finkelstein, R.; Gampala, S.S.; Lynch, T.J.; Thomas, T.L.; Rock, C.D. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol 2005, 59, 253–267. [Google Scholar]
- Bensmihen, S.; Rippa, S.; Lambert, G.; Jublot, D.; Pautot, V.; Granier, F.; Giraudat, J.; Parcy, F. The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 2002, 14, 1391–1403. [Google Scholar]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 2002, 32, 317–328. [Google Scholar]
- Liu, J.X.; Srivastava, R.; Che, P.; Howell, S.H. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J 2007, 51, 897–909. [Google Scholar]
- Yang, O.; Popova, O.V.; Suthoff, U.; Luking, I.; Dietz, K.J.; Golldack, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar]
- Amir, H.M.; Lee, Y.; Cho, J.I.; Ahn, C.H.; Lee, S.K.; Jeon, J.S.; Kang, H.; Lee, C.H.; An, G.; Park, P.B. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol 2010, 72, 557–566. [Google Scholar]
- Hsieh, T.H.; Li, C.W.; Su, R.C.; Cheng, C.P.; Tsai, Y.C.; Chan, M.T. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 2010, 231, 1459–1473. [Google Scholar]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 2008, 148, 1938–1952. [Google Scholar]
- Tang, N.; Zhang, H.; Li, X.; Xiao, J.; Xiong, L. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol 2012, 158, 1755–1768. [Google Scholar]
- Gao, S.Q.; Chen, M.; Xu, Z.S.; Zhao, C.P.; Li, L.; Xu, H.J.; Tang, Y.M.; Zhao, X.; Ma, Y.Z. The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol. Biol 2011, 75, 537–553. [Google Scholar]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar]
- Haake, V.; Cook, D.; Riechmann, J.L.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 2002, 130, 639–648. [Google Scholar]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J 2008, 56, 613–626. [Google Scholar]
- Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Seki, M.; Miura, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol. Biol 2000, 42, 657–665. [Google Scholar]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci USA 2006, 103, 18822–18827. [Google Scholar]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, G.; Vierling, E.; von Koskull-Doring, P. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J 2008, 53, 264–274. [Google Scholar]
- Chen, H.; Hwang, J.E.; Lim, C.J.; Kim, D.Y.; Lee, S.Y.; Lim, C.O. Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem. Biophys. Res. Commun 2010, 401, 238–244. [Google Scholar]
- Lee, S.J.; Kang, J.Y.; Park, H.J.; Kim, M.D.; Bae, M.S.; Choi, H.I.; Kim, S.Y. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol 2010, 153, 716–727. [Google Scholar]
- Reeves, W.M.; Lynch, T.J.; Mobin, R.; Finkelstein, R.R. Direct targets of the transcription factors ABA-Insensitive(ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol. Biol 2011, 75, 347–363. [Google Scholar]
- Dong, C.J.; Liu, J.Y. The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol 2010, 10, 47. [Google Scholar]
- Krishnaswamy, S.; Verma, S.; Rahman, M.H.; Kav, N.N. Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol. Biol 2011, 75, 107–127. [Google Scholar]
- Pandey, G.K.; Grant, J.J.; Cheong, Y.H.; Kim, B.G.; Li, L.; Luan, S. ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 2005, 139, 1185–1193. [Google Scholar]
- Song, C.P.; Agarwal, M.; Ohta, M.; Guo, Y.; Halfter, U.; Wang, P.; Zhu, J.K. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 2005, 17, 2384–2396. [Google Scholar]
- Yang, S.; Vanderbeld, B.; Wan, J.; Huang, Y. Narrowing down the targets: Towards successful genetic engineering of drought-tolerant crops. Mol. Plant 2010, 3, 469–490. [Google Scholar]
- Xiao, B.Z.; Chen, X.; Xiang, C.B.; Tang, N.; Zhang, Q.F.; Xiong, L.Z. Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol. Plant 2009, 2, 73–83. [Google Scholar]
- Karaba, A.; Dixit, S.; Greco, R.; Aharoni, A.; Trijatmiko, K.R.; Marsch-Martinez, N.; Krishnan, A.; Nataraja, K.N.; Udayakumar, M.; Pereira, A. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc. Natl. Acad. Sci. USA 2007, 104, 15270–15275. [Google Scholar]
- Cui, M.; Zhang, W.; Zhang, Q.; Xu, Z.; Zhu, Z.; Duan, F.; Wu, R. Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem 2011, 49, 1384–1391. [Google Scholar]
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’Shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J 2010, 426, 183–196. [Google Scholar]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci 2005, 10, 79–87. [Google Scholar]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Osakabe, Y.; Qin, F.; Simpson, S.D.; Maruyama, K.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J 2007, 49, 46–63. [Google Scholar]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 2004, 39, 863–876. [Google Scholar]
- Lu, P.L.; Chen, N.Z.; An, R.; Su, Z.; Qi, B.S.; Ren, F.; Chen, J.; Wang, X.C. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol 2007, 63, 289–305. [Google Scholar]
- Wu, Y.; Deng, Z.; Lai, J.; Zhang, Y.; Yang, C.; Yin, B.; Zhao, Q.; Zhang, L.; Li, Y.; Yang, C.; et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 2009, 19, 1279–1290. [Google Scholar]
- Bu, Q.; Jiang, H.; Li, C.B.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Li, C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 2008, 18, 756–767. [Google Scholar]
- Jiang, H.; Li, H.; Bu, Q.; Li, C. The RHA2a-interacting proteins ANAC019 and ANAC055 may play a dual role in regulating ABA response and jasmonate response. Plant Signal. Behav 2009, 4, 464–466. [Google Scholar]
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 2005, 44, 903–916. [Google Scholar]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Kohler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 2010, 62, 250–264. [Google Scholar]
- Yang, S.D.; Seo, P.J.; Yoon, H.K.; Park, C.M. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar]
- Yamaguchi, M.; Ohtani, M.; Mitsuda, N.; Kubo, M.; Ohme-Takagi, M.; Fukuda, H.; Demura, T. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 2010, 22, 1249–1263. [Google Scholar]
- Seo, P.J.; Kim, M.J.; Park, J.Y.; Kim, S.Y.; Jeon, J.; Lee, Y.H.; Kim, J.; Park, C.M. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J 2010, 61, 661–671. [Google Scholar]
- Seo, P.J.; Park, C.M. A membrane-bound NAC transcription factor as an integrator of biotic and abiotic stress signals. Plant Signal. Behav 2010, 5, 481–483. [Google Scholar]
- Kim, S.G.; Lee, A.K.; Yoon, H.K.; Park, C.M. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J 2008, 55, 77–88. [Google Scholar]
- Yoon, H.K.; Kim, S.G.; Kim, S.Y.; Park, C.M. Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol. Cells 2008, 25, 438–445. [Google Scholar]
- Lee, S.; Seo, P.J.; Lee, H.J.; Park, C.M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 2012, 70, 831–844. [Google Scholar]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol 2008, 67, 169–181. [Google Scholar]
- Nakashima, K.; Tran, L.S.; Van, N.D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 2007, 51, 617–630. [Google Scholar]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Nakashima, K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 2010, 284, 173–183. [Google Scholar]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Do, C.Y.; Kim, M.; Reuzeau, C.; Kim, J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 2010, 153, 185–197. [Google Scholar]
- Zheng, X.; Chen, B.; Lu, G.; Han, B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun 2009, 379, 985–989. [Google Scholar]
- Xue, G.P.; Way, H.M.; Richardson, T.; Drenth, J.; Joyce, P.A.; McIntyre, C.L. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 2011, 4, 697–712. [Google Scholar]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K.; et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J 2011, 68, 302–313. [Google Scholar]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci 2000, 5, 199–206. [Google Scholar]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol 2009, 69, 91–105. [Google Scholar]
- Li, S.; Fu, Q.; Chen, L.; Huang, W.; Yu, D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 2011, 233, 1237–1252. [Google Scholar]
- Andreasson, E.; Jenkins, T.; Brodersen, P.; Thorgrimsen, S.; Petersen, N.H.; Zhu, S.; Qiu, J.L.; Micheelsen, P.; Rocher, A.; Petersen, M.; et al. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 2005, 24, 2579–2589. [Google Scholar]
- Li, S.; Zhou, X.; Chen, L.; Huang, W.; Yu, D. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells 2010, 29, 475–483. [Google Scholar]
- Zou, C.; Jiang, W.; Yu, D. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J. Exp. Bot 2010, 61, 3901–3914. [Google Scholar]
- Jiang, Y.; Liang, G.; Yu, D. Activated Expression of WRKY57 Confers Drought Tolerance in Arabidopsis. Mol. Plant 2012, 5, 1375–1388. [Google Scholar]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J 2010, 63, 417–429. [Google Scholar]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 2010, 10, 281. [Google Scholar]
- Shang, Y.; Yan, L.; Liu, Z.Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.Q.; Wang, X.F.; Du, S.Y.; Jiang, T.; et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar]
- Liu, Z.Q.; Yan, L.; Wu, Z.; Mei, C.; Lu, K.; Yu, Y.T.; Liang, S.; Zhang, X.F.; Wang, X.F.; Zhang, D.P. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J. Exp. Bot 2012, 63, 6371–6392. [Google Scholar]
- Jiang, W.; Yu, D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol 2009, 9, 96. [Google Scholar]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep 2009, 28, 21–30. [Google Scholar]
- Tao, Z.; Kou, Y.; Liu, H.; Li, X.; Xiao, J.; Wang, S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot 2011, 62, 4863–4874. [Google Scholar]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J 2008, 6, 486–503. [Google Scholar]
- Englbrecht, C.C.; Schoof, H.; Bohm, S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 2004, 5, 39. [Google Scholar]
- Ciftci-Yilmaz, S.; Morsy, M.R.; Song, L.; Coutu, A.; Krizek, B.A.; Lewis, M.W.; Warren, D.; Cushman, J.; Connolly, E.L.; Mittler, R. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J. Biol. Chem 2007, 282, 9260–9268. [Google Scholar]
- Lee, H.; Guo, Y.; Ohta, M.; Xiong, L.; Stevenson, B.; Zhu, J.K. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J 2002, 21, 2692–2702. [Google Scholar]
- Mittler, R.; Kim, Y.; Song, L.; Coutu, J.; Coutu, A.; Ciftci-Yilmaz, S.; Lee, H.; Stevenson, B.; Zhu, J.K. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 2006, 580, 6537–6542. [Google Scholar]
- Vogel, J.T.; Zarka, D.G.; Van Buskirk, H.A.; Fowler, S.G.; Thomashow, M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 2005, 41, 195–211. [Google Scholar]
- Kodaira, K.S.; Qin, F.; Tran, L.S.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol 2011, 157, 742–756. [Google Scholar]
- Jiang, C.J.; Aono, M.; Tamaoki, M.; Maeda, S.; Sugano, S.; Mori, M.; Takatsuji, H. SAZ, a new SUPERMAN-like protein, negatively regulates a subset of ABA-responsive genes in Arabidopsis. Mol. Genet. Genomics 2008, 279, 183–192. [Google Scholar]
- Huang, X.Y.; Chao, D.Y.; Gao, J.P.; Zhu, M.Z.; Shi, M.; Lin, H.X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 2009, 23, 1805–1817. [Google Scholar]
- Xu, D.Q.; Huang, J.; Guo, S.Q.; Yang, X.; Bao, Y.M.; Tang, H.J.; Zhang, H.S. Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.). FEBS Lett 2008, 582, 1037–1043. [Google Scholar]
- Huang, J.; Sun, S.; Xu, D.; Lan, H.; Sun, H.; Wang, Z.; Bao, Y.; Wang, J.; Tang, H.; Zhang, H. A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Mol. Biol 2012, 80, 337–350. [Google Scholar]
- Cominelli, E.; Galbiati, M.; Vavasseur, A.; Conti, L.; Sala, T.; Vuylsteke, M.; Leonhardt, N.; Dellaporta, S.L.; Tonelli, C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol 2005, 15, 1196–1200. [Google Scholar]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 2008, 146, 623–635. [Google Scholar]
- Jung, C.; Shim, J.S.; Seo, J.S.; Lee, H.Y.; Kim, C.H.; Choi, Y.D.; Cheong, J.J. Non-specific phytohormonal induction of AtMYB44 and suppression of jasmonate-responsive gene activation in Arabidopsis thaliana. Mol. Cells 2010, 29, 71–86. [Google Scholar]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 2009, 151, 275–289. [Google Scholar]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem 2006, 281, 37636–37645. [Google Scholar]
- Ding, Z.; Li, S.; An, X.; Liu, X.; Qin, H.; Wang, D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genomics 2009, 36, 17–29. [Google Scholar]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot 2012, 63, 2541–2556. [Google Scholar]
- Ma, Q.; Dai, X.; Xu, Y.; Guo, J.; Liu, Y.; Chen, N.; Xiao, J.; Zhang, D.; Xu, Z.; Zhang, X.; et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 2009, 150, 244–256. [Google Scholar]
- Zhang, Z.; Liu, X.; Wang, X.; Zhou, M.; Zhou, X.; Ye, X.; Wei, X. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol 2012, 196, 1155–1170. [Google Scholar]
- Bailey, P.C.; Martin, C.; Toledo-Ortiz, G.; Quail, P.H.; Huq, E.; Heim, M.A.; Jakoby, M.; Werber, M.; Weisshaar, B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 2003, 15, 2497–2502. [Google Scholar]
- Kazan, K.; Manners, J.M. MYC2: The Master in Action. Mol. Plant 2012. [Google Scholar] [CrossRef]
- Li, H.; Sun, J.; Xu, Y.; Jiang, H.; Wu, X.; Li, C. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol. Biol 2007, 65, 655–665. [Google Scholar]
- Jiang, Y.; Yang, B.; Deyholos, M.K. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Genet. Genomics 2009, 282, 503–516. [Google Scholar]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 2003, 17, 1043–1054. [Google Scholar]
- Zhu, Y.; Yang, H.; Mang, H.G.; Hua, J. Induction of BAP1 by a moderate decrease in temperature is mediated by ICE1 in Arabidopsis. Plant Physiol 2011, 155, 580–588. [Google Scholar]
- Kim, J.S.; Mizoi, J.; Kidokoro, S.; Maruyama, K.; Nakajima, J.; Nakashima, K.; Mitsuda, N.; Takiguchi, Y.; Ohme-Takagi, M.; Kondou, Y.; et al. Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 2012, 24, 3393–3405. [Google Scholar]
- Yoshida, T.; Ohama, N.; Nakajima, J.; Kidokoro, S.; Mizoi, J.; Nakashima, K.; Maruyama, K.; Kim, J.M.; Seki, M.; Todaka, D.; et al. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genomics 2011, 286, 321–332. [Google Scholar]
- Park, J.; Lee, N.; Kim, W.; Lim, S.; Choi, G. ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 2011, 23, 1404–1415. [Google Scholar]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 2005, 10, 88–94. [Google Scholar]
- Tompa, P.; Kovacs, D. Intrinsically disordered chaperones in plants and animals. Biochem. Cell Biol 2010, 88, 167–174. [Google Scholar]
- Luscombe, N.M.; Thornton, J.M. Protein-DNA interactions: Amino acid conservation and the effects of mutations on binding specificity. J. Mol. Biol 2002, 320, 991–1009. [Google Scholar]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat Rev Mol. Cell Biol 2005, 6, 197–208. [Google Scholar]
- Kjaersgaard, T.; Jensen, M.K.; Christiansen, M.W.; Gregersen, P.; Kragelund, B.B.; Skriver, K. Senescence-associated barley NAC (NAM, ATAF1,2, CUC) transcription factor interacts with radical-induced cell death 1 through a disordered regulatory domain. J. Biol. Chem 2011, 286, 35418–35429. [Google Scholar]
- Kragelund, B.B.; Jensen, M.K.; Skriver, K. Order by disorder in plant signaling. Trends Plant Sci 2012, 17, 625–632. [Google Scholar]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar]
- Schumacher, M.A.; Goodman, R.H.; Brennan, R.G. The structure of a CREB bZIP.somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J. Biol. Chem 2000, 275, 35242–35247. [Google Scholar]
- Skriver, K.; Olsen, F.L.; Rogers, J.C.; Mundy, J. cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc. Natl. Acad. Sci. USA 1991, 88, 7266–7270. [Google Scholar]
- Nakamura, S.; Lynch, T.J.; Finkelstein, R.R. Physical interactions between ABA response loci of Arabidopsis. Plant J 2001, 26, 627–635. [Google Scholar]
- Yamasaki, K.; Kigawa, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci 2012. [Google Scholar] [CrossRef]
- Allen, M.D.; Yamasaki, K.; Ohme-Takagi, M.; Tateno, M.; Suzuki, M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J 1998, 17, 5484–5496. [Google Scholar]
- Canella, D.; Gilmour, S.J.; Kuhn, L.A.; Thomashow, M.F. DNA binding by the Arabidopsis CBF1 transcription factor requires the PKKP/RAGRxKFxETRHP signature sequence. Biochim. Biophys. Acta 2010, 1799, 454–462. [Google Scholar]
- Nole-Wilson, S.; Krizek, B.A. DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res 2000, 28, 4076–4082. [Google Scholar]
- Kagaya, Y.; Ohmiya, K.; Hattori, T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 1999, 27, 470–478. [Google Scholar]
- Blomberg, J.; Aguilar, X.; Brannstrom, K.; Rautio, L.; Olofsson, A.; Wittung-Stafshede, P.; Bjorklund, S. Interactions between DNA, transcriptional regulator Dreb2a and the Med25 mediator subunit from Arabidopsis thaliana involve conformational changes. Nucleic Acids Res 2012, 40, 5938–5950. [Google Scholar]
- Ernst, H.A.; Olsen, A.N.; Larsen, S.; Lo, L.L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 2004, 5, 297–303. [Google Scholar]
- Chen, Q.; Wang, Q.; Xiong, L.; Lou, Z. A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell 2011, 2, 55–63. [Google Scholar]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. DNA-binding specificity and molecular finctions of NAC transcription factros. Plant Sci 2005, 169, 785–797. [Google Scholar]
- Welner, D.H.; Lindemose, S.; Grossmann, J.G.; Mollegaard, N.E.; Olsen, A.N.; Helgstrand, C.; Skriver, K.; Lo, L.L. DNA binding by the plant-specific NAC transcription factors in crystal and solution: A firm link to WRKY and GCM transcription factors. Biochem J 2012, 444, 395–404. [Google Scholar]
- Cohen, S.X.; Moulin, M.; Hashemolhosseini, S.; Kilian, K.; Wegner, M.; Muller, C.W. Structure of the GCM domain-DNA complex: A DNA-binding domain with a novel fold and mode of target site recognition. EMBO J 2003, 22, 1835–1845. [Google Scholar]
- Yamasaki, K.; Kigawa, T.; Watanabe, S.; Inoue, M.; Yamasaki, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. Structural basis for sequence-specific DNA recognition by an Arabidopsis WRKY transcription factor. J. Biol. Chem 2012, 287, 7683–7691. [Google Scholar]
- Duan, M.R.; Nan, J.; Liang, Y.H.; Mao, P.; Lu, L.; Li, L.; Wei, C.; Lai, L.; Li, Y.; Su, X.D. DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res 2007, 35, 1145–1154. [Google Scholar]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Tomo, Y.; et al. Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell 2005, 17, 944–956. [Google Scholar]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci 2010, 15, 247–258. [Google Scholar]
- Maeo, K.; Hayashi, S.; Kojima-Suzuki, H.; Morikami, A.; Nakamura, K. Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci. Biotechnol. Biochem 2001, 65, 2428–2436. [Google Scholar]
- Pavletich, N.P.; Pabo, C.O. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 A. Science 1991, 252, 809–817. [Google Scholar]
- Kubo, K.; Sakamoto, A.; Kobayashi, A.; Rybka, Z.; Kanno, Y.; Nakagawa, H.; Takatsuji, H. Cys2/His2 zinc-finger protein family of petunia: Evolution and general mechanism of target-sequence recognition. Nucleic Acids Res 1998, 26, 608–615. [Google Scholar]
- Ogata, K.; Kanei-Ishii, C.; Sasaki, M.; Hatanaka, H.; Nagadoi, A.; Enari, M.; Nakamura, H.; Nishimura, Y.; Ishii, S.; Sarai, A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol 1996, 3, 178–187. [Google Scholar]
- Romero, I.; Fuertes, A.; Benito, M.J.; Malpica, J.M.; Leyva, A.; Paz-Ares, J. More than 80R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J 1998, 14, 273–284. [Google Scholar]
- Hanaoka, S.; Nagadoi, A.; Nishimura, Y. Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci 2005, 14, 119–130. [Google Scholar]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar]
- Ma, P.C.; Rould, M.A.; Weintraub, H.; Pabo, C.O. Crystal structure of MyoD bHLH domain-DNA complex: Perspectives on DNA recognition and implications for transcriptional activation. Cell 1994, 77, 451–459. [Google Scholar]
- Fuxreiter, M.; Simon, I.; Bondos, S. Dynamic protein-DNA recognition: Beyond what can be seen. Trends Biochem. Sci 2001, 36, 415–423. [Google Scholar]
- Guo, X.; Bulyk, M.L.; Hartemink, A.J. Intrinsic disorder within and flanking the DNA-binding domains of human transcription factors. Pac. Symp. Biocomput. 2012, 104–115. [Google Scholar]
- Fuxreiter, M.; Tompa, P.; Simon, I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics 2007, 23, 950–956. [Google Scholar]
- Han, J.D.; Bertin, N.; Hao, T.; Goldberg, D.S.; Berriz, G.F.; Zhang, L.V.; Dupuy, D.; Walhout, A.J.; Cusick, M.E.; Roth, F.P.; et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 2004, 430, 88–93. [Google Scholar]
- Han, S.K.; Sang, Y.; Rodrigues, A.; Wu, M.F.; Rodriguez, P.L.; Wagner, D. The SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA Represses Abscisic Acid Responses in the Absence of the Stress Stimulus in Arabidopsis. Plant Cell 2012, 24, 4892–4906. [Google Scholar]
- Li, B.; Qin, Y.; Duan, H.; Yin, W.; Xia, X. Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J. Exp. Bot 2011, 62, 3765–3779. [Google Scholar]
- Vainonen, J.P.; Jaspers, P.; Wrzaczek, M.; Lamminmaki, A.; Reddy, R.A.; Vaahtera, L.; Brosche, M.; Kangasjarvi, J. RCD1-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem. J 2012, 442, 573–581. [Google Scholar]
- Kao, H.L.; Gunsalus, K.C. Browsing multidimensional molecular networks with the generic network browser (N-Browse). Curr. Protoc. Bioinform. 2008. [Google Scholar] [CrossRef]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 2009, 50, 1345–1363. [Google Scholar]
- Garcia, M.E.; Lynch, T.; Peeters, J.; Snowden, C.; Finkelstein, R. A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol 2008, 67, 643–658. [Google Scholar]
- Lopez-Molina, L.; Mongrand, S.; Kinoshita, N.; Chua, N.H. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 2003, 17, 410–418. [Google Scholar]
- Liu, H.; Stone, S.L. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell. 2010, 22, 2630–2641. [Google Scholar]
- Stone, S.L.; Williams, L.A.; Farmer, L.M.; Vierstra, R.D.; Callis, J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 2006, 18, 3415–3428. [Google Scholar]
- Smalle, J.; Kurepa, J.; Yang, P.; Emborg, T.J.; Babiychuk, E.; Kushnir, S.; Vierstra, R.D. The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 2003, 15, 965–980. [Google Scholar]
- Miura, K.; Hasegawa, P.M. Sumoylation and abscisic acid signaling. Plant Signal. Behav 2009, 4, 1176–1178. [Google Scholar]
- Chen, R.; Jiang, H.; Li, L.; Zhai, Q.; Qi, L.; Zhou, W.; Liu, X.; Li, H.; Zheng, W.; Sun, J.; et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 2012, 24, 2898–2916. [Google Scholar]
- Qin, F.; Sakuma, Y.; Tran, L.S.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.; et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar]
- Jaspers, P.; Blomster, T.; Brosche, M.; Salojarvi, J.; Ahlfors, R.; Vainonen, J.P.; Reddy, R.A.; Immink, R.; Angenent, G.; Turck, F.; et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J 2009, 60, 268–279. [Google Scholar]
- Greve, K.; La Cour, T.; Jensen, M.K.; Poulsen, F.M.; Skriver, K. Interactions between plant RING-H2 and plant-specific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochem J 2003, 371, 97–108. [Google Scholar]
- Xie, Q.; Guo, H.S.; Dallman, G.; Fang, S.; Weissman, A.M.; Chua, N.H. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 2002, 419, 167–170. [Google Scholar]
- Bu, Q.; Li, H.; Zhao, Q.; Jiang, H.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Wang, D.; et al. The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol 2009, 150, 463–481. [Google Scholar]
- Bang, W.Y.; Kim, S.W.; Jeong, I.S.; Koiwa, H.; Bahk, J.D. The C-terminal region (640–967) of Arabidopsis CPL1 interacts with the abiotic stress- and ABA-responsive transcription factors. Biochem. Biophys. Res. Commun 2008, 372, 907–912. [Google Scholar]
- Xu, X.; Chen, C.; Fan, B.; Chen, Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 2006, 18, 1310–1326. [Google Scholar]
- Davletova, S.; Rizhsky, L.; Liang, H.; Shengqiang, Z.; Oliver, D.J.; Coutu, J.; Shulaev, V.; Schlauch, K.; Mittler, R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 2005, 17, 268–281. [Google Scholar]
- Pnueli, L.; Liang, H.; Rozenberg, M.; Mittler, R. Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 2003, 34, 187–203. [Google Scholar]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar]
- Dong, C.H.; Agarwal, M.; Zhang, Y.; Xie, Q.; Zhu, J.K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 2006, 103, 8281–8286. [Google Scholar]
- Miura, K.; Ohta, M.; Nakazawa, M.; Ono, M.; Hasegawa, P.M. ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance. Plant J 2011, 67, 269–279. [Google Scholar] [Green Version]
- Miura, K.; Hasegawa, P.M. Regulation of cold signaling by sumoylation of ICE1. Plant Signal. Behav 2008, 3, 52–53. [Google Scholar]
- Miura, K.; Jin, J.B.; Lee, J.; Yoo, C.Y.; Stirm, V.; Miura, T.; Ashworth, E.N.; Bressan, R.A.; Yun, D.J.; Hasegawa, P.M. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 2007, 19, 1403–1414. [Google Scholar]
- Chen, C.C.; Liang, C.S.; Kao, A.L.; Yang, C.C. HHP1, a novel signalling component in the cross-talk between the cold and osmotic signalling pathways in Arabidopsis. J. Exp. Bot 2010, 61, 3305–3320. [Google Scholar]
- Kanaoka, M.M.; Pillitteri, L.J.; Fujii, H.; Yoshida, Y.; Bogenschutz, N.L.; Takabayashi, J.; Zhu, J.K.; Torii, K.U. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell 2008, 20, 1775–1785. [Google Scholar]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol 2012, 196, 13–28. [Google Scholar]
- Castro, P.H.; Tavares, R.M.; Bejarano, E.R.; Azevedo, H. SUMO, a heavyweight player in plant abiotic stress responses. Cell. Mol. Life. Sci 2012, 69, 3269–3283. [Google Scholar]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J 2005, 272, 5129–5148. [Google Scholar]
- Oldfield, C.J.; Meng, J.; Yang, J.Y.; Yang, M.Q.; Uversky, V.N.; Dunker, A.K. Flexible nets: Disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 2008, 9. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lindemose, S.; O'Shea, C.; Jensen, M.K.; Skriver, K. Structure, Function and Networks of Transcription Factors Involved in Abiotic Stress Responses. Int. J. Mol. Sci. 2013, 14, 5842-5878. https://doi.org/10.3390/ijms14035842
Lindemose S, O'Shea C, Jensen MK, Skriver K. Structure, Function and Networks of Transcription Factors Involved in Abiotic Stress Responses. International Journal of Molecular Sciences. 2013; 14(3):5842-5878. https://doi.org/10.3390/ijms14035842
Chicago/Turabian StyleLindemose, Søren, Charlotte O'Shea, Michael Krogh Jensen, and Karen Skriver. 2013. "Structure, Function and Networks of Transcription Factors Involved in Abiotic Stress Responses" International Journal of Molecular Sciences 14, no. 3: 5842-5878. https://doi.org/10.3390/ijms14035842




