Structural Biology of a Major Signaling Network that Regulates Plant Abiotic Stress: The CBL-CIPK Mediated Pathway
Abstract
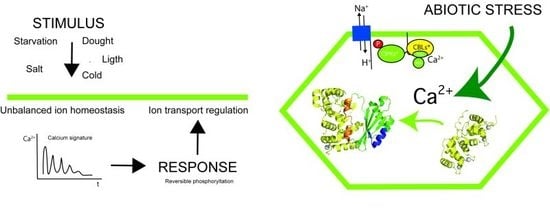
:1. Introduction
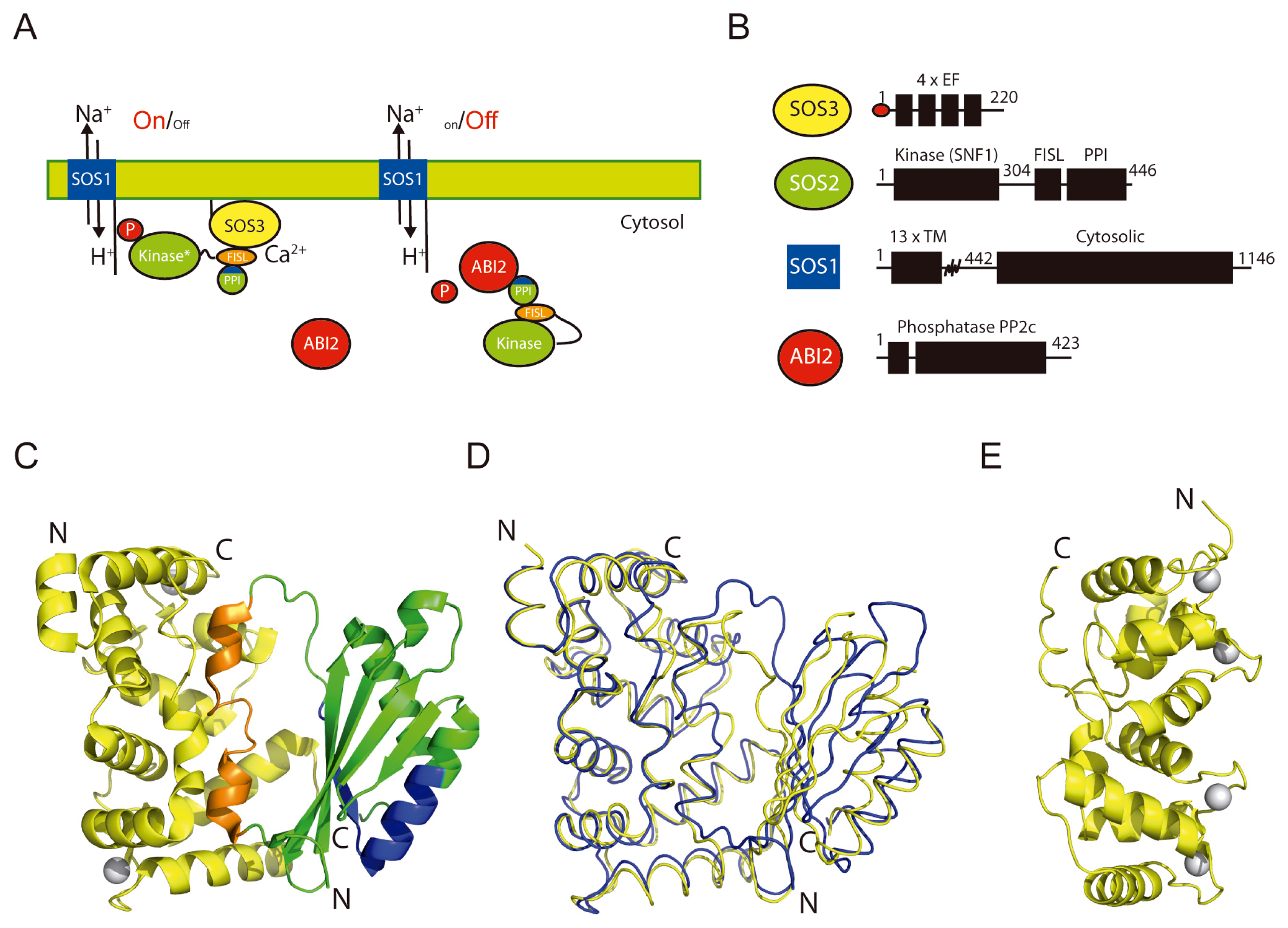
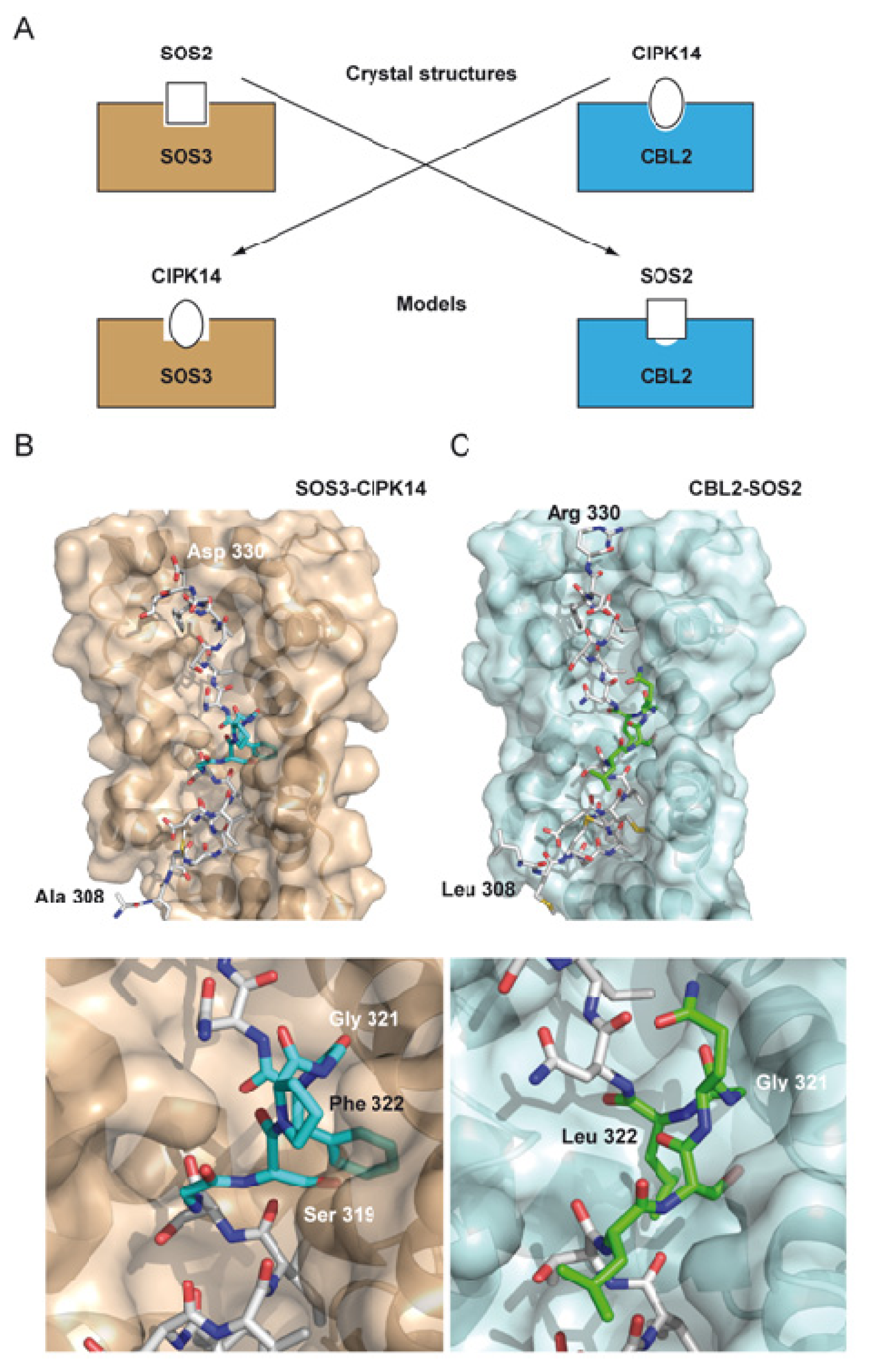
2. The Overall Architecture of the CBL-CIPK Complexes Provides the Structural Basis of Key Aspects of the Regulation of the Network
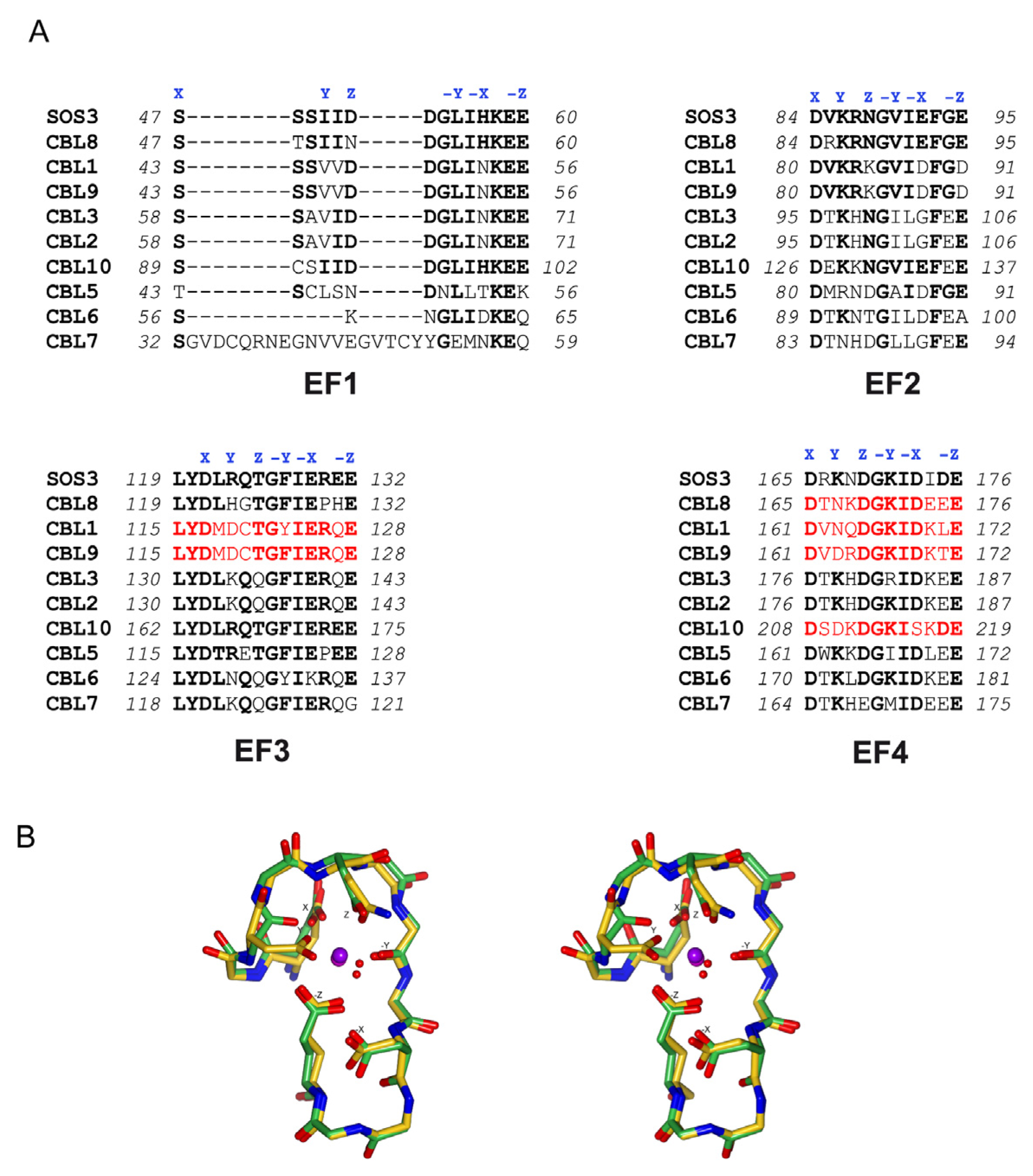
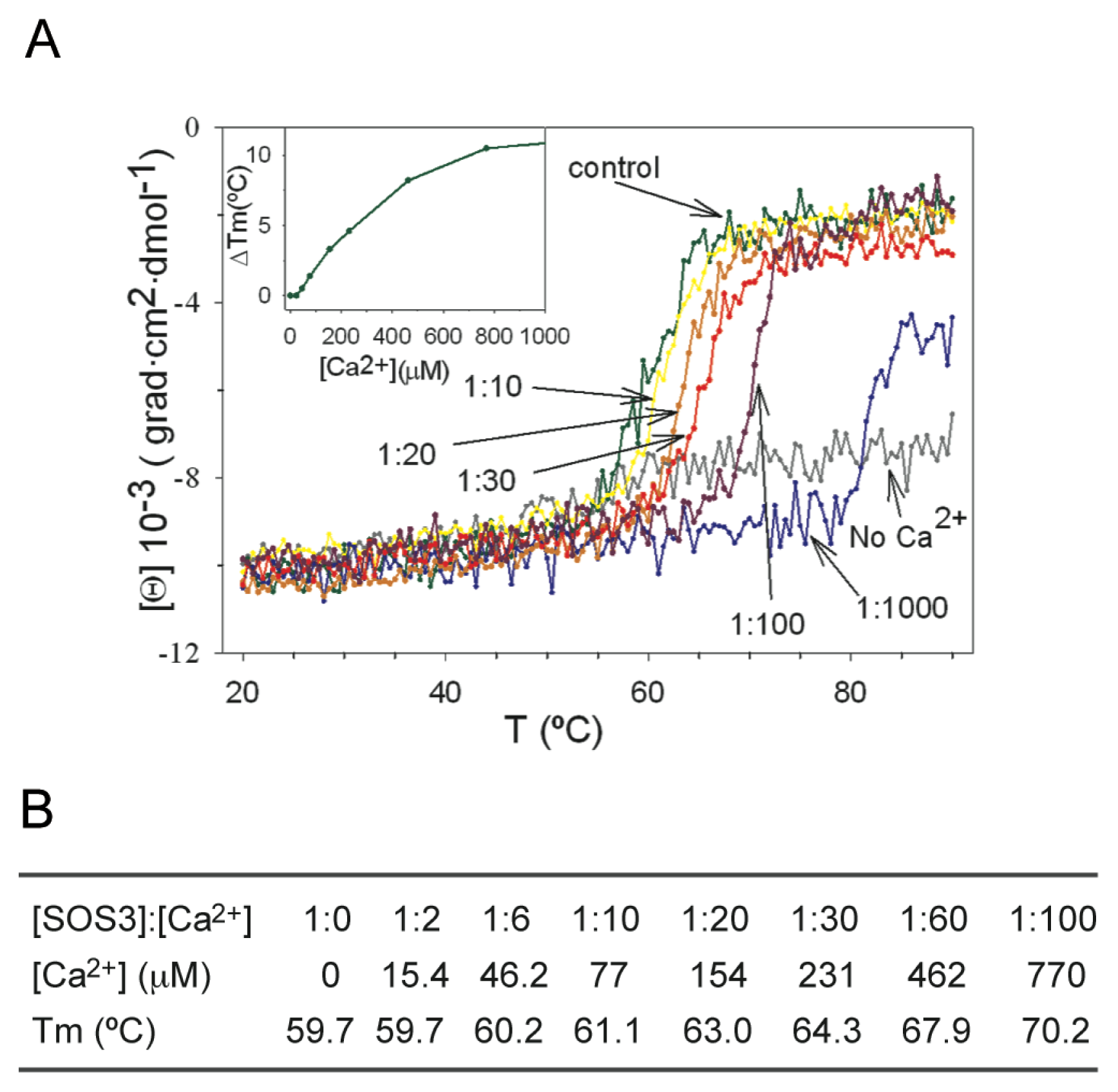
3. Decoding the Calcium Signal
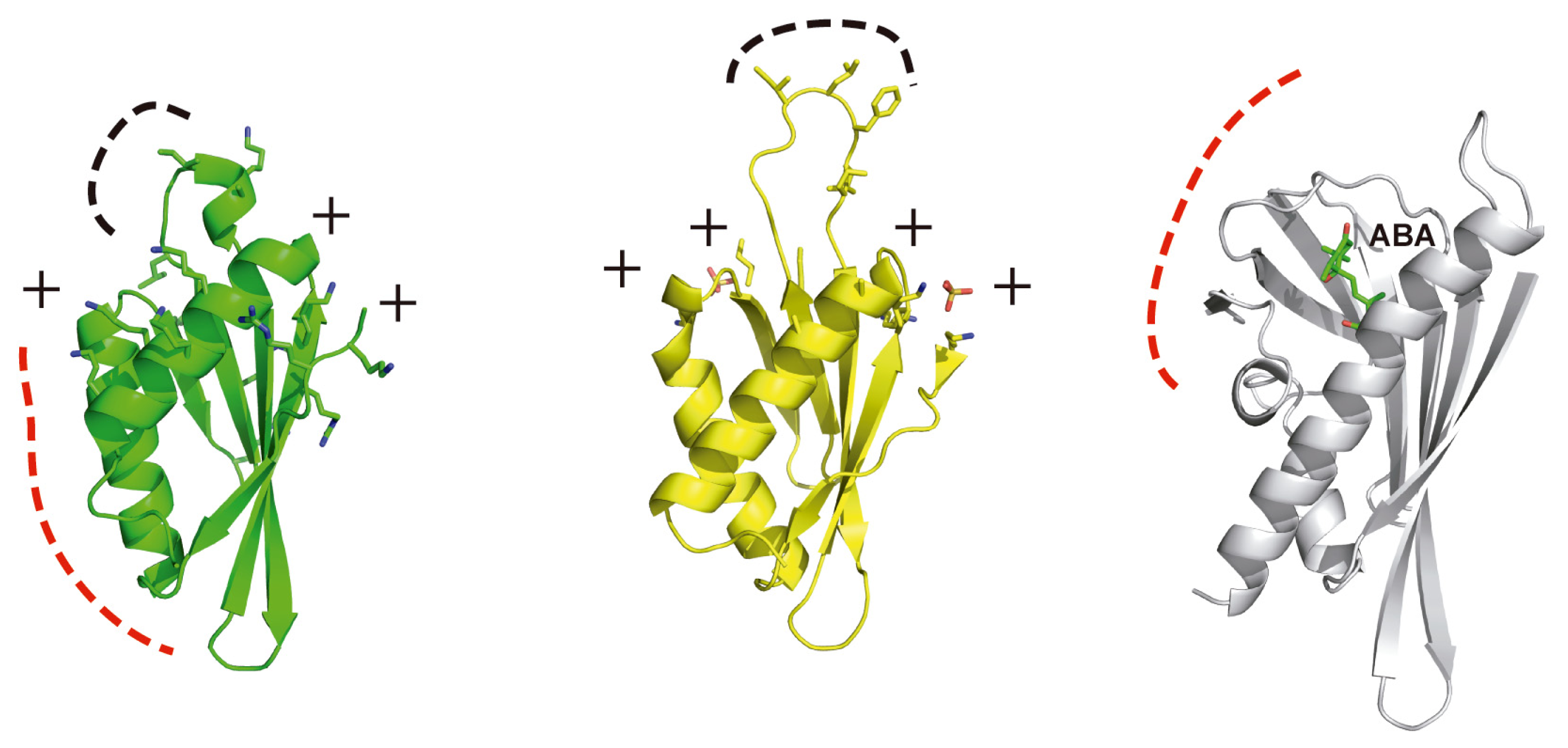
4. Structural Basis of Localization of CBL Proteins and CIPKs
5. Final Remarks
Acknowledgments
Conflict of Interest
References
- Jura, N.; Zhang, X.; Endres, N.F.; Seeliger, M.A.; Schindler, T.; Kuriyan, J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol. Cell 2011, 42, 9–22. [Google Scholar]
- Shi, J.; Kim, K.N.; Ritz, O.; Albrecht, V.; Gupta, R.; Harter, K.; Luan, S.; Kudla, J. Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 1999, 11, 2393–2405. [Google Scholar]
- Albrecht, V.; Ritz, O.; Linder, S.; Harter, K.; Kudla, J. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J 2001, 20, 1051–1063. [Google Scholar]
- Drerup, M.M.; Schlucking, K.; Hashimoto, K.; Manishankar, P.; Steinhorst, L.; Kuchitsu, K.; Kudla, J. The calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant 2013. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar]
- Quintero, F.J.; Martinez-Atienza, J.; Villalta, I.; Jiang, X.; Kim, W.Y.; Ali, Z.; Fujii, H.; Mendoza, I.; Yun, D.J.; Zhu, J.K.; et al. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 2011, 108, 2611–2616. [Google Scholar]
- Xu, J.; Li, H.D.; Chen, L.Q.; Wang, Y.; Liu, L.L.; He, L.; Wu, W.H. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 2006, 125, 1347–1360. [Google Scholar]
- Held, K.; Pascaud, F.; Eckert, C.; Gajdanowicz, P.; Hashimoto, K.; Corratge-Faillie, C.; Offenborn, J.N.; Lacombe, B.; Dreyer, I.; Thibaud, J.B.; et al. Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res 2011, 21, 1116–1130. [Google Scholar]
- Li, L.; Kim, B.G.; Cheong, Y.H.; Pandey, G.K.; Luan, S. A Ca(2)+ signaling pathway regulates a K(+) channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 12625–12630. [Google Scholar]
- Lee, S.C.; Lan, W.Z.; Kim, B.G.; Li, L.; Cheong, Y.H.; Pandey, G.K.; Lu, G.; Buchanan, B.B.; Luan, S. A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl. Acad. Sci. USA 2007, 104, 15959–15964. [Google Scholar]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar]
- Kim, B.G.; Waadt, R.; Cheong, Y.H.; Pandey, G.K.; Dominguez-Solis, J.R.; Schultke, S.; Lee, S.C.; Kudla, J.; Luan, S. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J 2007, 52, 473–484. [Google Scholar]
- Fuglsang, A.T.; Guo, Y.; Cuin, T.A.; Qiu, Q.; Song, C.; Kristiansen, K.A.; Bych, K.; Schulz, A.; Shabala, S.; Schumaker, K.S.; et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 2007, 19, 1617–1634. [Google Scholar]
- Halfter, U.; Ishitani, M.; Zhu, J.K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar]
- Liu, J.; Ishitani, M.; Halfter, U.; Kim, C.S.; Zhu, J.K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 2000, 97, 3730–3734. [Google Scholar]
- Zhu, J.K. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 2000, 124, 941–948. [Google Scholar]
- Guo, Y.; Halfter, U.; Ishitani, M.; Zhu, J.K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 2001, 13, 1383–1400. [Google Scholar]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol 2003, 6, 441–445. [Google Scholar]
- Guo, Y.; Qiu, Q.S.; Quintero, F.J.; Pardo, J.M.; Ohta, M.; Zhang, C.; Schumaker, K.S.; Zhu, J.K. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 2004, 16, 435–449. [Google Scholar]
- Liu, J.; Zhu, J.K. A calcium sensor homolog required for plant salt tolerance. Science 1998, 280, 1943–1945. [Google Scholar]
- Nunez-Ramirez, R.; Sanchez-Barrena, M.J.; Villalta, I.; Vega, J.F.; Pardo, J.M.; Quintero, F.J.; Martinez-Salazar, J.; Albert, A. Structural insights on the plant Salt-Overly-Sensitive 1 (SOS1) Na(+)/H(+) antiporter. J. Mol. Biol 2012, 424, 283–294. [Google Scholar]
- Akaboshi, M.; Hashimoto, H.; Ishida, H.; Saijo, S.; Koizumi, N.; Sato, M.; Shimizu, T. The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14. J. Mol. Biol 2008, 377, 246–257. [Google Scholar]
- Sanchez-Barrena, M.J.; Fujii, H.; Angulo, I.; Martinez-Ripoll, M.; Zhu, J.K.; Albert, A. The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol. Cell 2007, 26, 427–435. [Google Scholar]
- Sanchez-Barrena, M.J.; Moreno-Perez, S.; Angulo, I.; Martinez-Ripoll, M.; Albert, A. The complex between SOS3 and SOS2 regulatory domain from Arabidopsis thaliana: Cloning, expression, purification, crystallization and preliminary X-ray analysis. Acta crystallogr. Sect. F 2007, 63, 568–570. [Google Scholar]
- Yunta, C.; Martinez-Ripoll, M.; Zhu, J.K.; Albert, A. The structure of Arabidopsis thaliana OST1 provides insights into the kinase regulation mechanism in response to osmotic stress. J. Mol. Biol 2011, 414, 135–144. [Google Scholar]
- Ohta, M.; Guo, Y.; Halfter, U.; Zhu, J.K. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 2003, 100, 11771–11776. [Google Scholar]
- Sanchez-Barrena, M.J.; Martinez-Ripoll, M.; Zhu, J.K.; Albert, A. SOS3 (salt overly sensitive 3) from Arabidopsis thaliana: Expression, purification, crystallization and preliminary X-ray analysis. Acta crystallogr. Sect. D 2004, 60, 1272–1274. [Google Scholar]
- Sanchez-Barrena, M.J.; Martinez-Ripoll, M.; Zhu, J.K.; Albert, A. The structure of the Arabidopsis thaliana SOS3: Molecular mechanism of sensing calcium for salt stress response. J. Mol. Biol 2005, 345, 1253–1264. [Google Scholar]
- Gong, D.; Guo, Y.; Schumaker, K.S.; Zhu, J.K. The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol 2004, 134, 919–926. [Google Scholar]
- Batistic, O.; Kudla, J. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 2004, 219, 915–924. [Google Scholar]
- Nagae, M.; Nozawa, A.; Koizumi, N.; Sano, H.; Hashimoto, H.; Sato, M.; Shimizu, T. The crystal structure of the novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. J. Biol. Chem 2003, 278, 42240–42246. [Google Scholar]
- De Diego, I.; Kuper, J.; Bakalova, N.; Kursula, P.; Wilmanns, M. Molecular basis of the death-associated protein kinase-calcium/calmodulin regulator complex. Sci. Signal. 2010, 3, ra6. [Google Scholar]
- Kissinger, C.R.; Parge, H.E.; Knighton, D.R.; Lewis, C.T.; Pelletier, L.A.; Tempczyk, A.; Kalish, V.J.; Tucker, K.D.; Showalter, R.E.; Moomaw, E.W.; et al. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature 1995, 378, 641–644. [Google Scholar]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar]
- Melcher, K.; Ng, L.M.; Zhou, X.E.; Soon, F.F.; Xu, Y.; Suino-Powell, K.M.; Park, S.Y.; Weiner, J.J.; Fujii, H.; Chinnusamy, V.; et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 2009, 462, 602–608. [Google Scholar]
- Miyazono, K.; Miyakawa, T.; Sawano, Y.; Kubota, K.; Kang, H.J.; Asano, A.; Miyauchi, Y.; Takahashi, M.; Zhi, Y.; Fujita, Y.; et al. Structural basis of abscisic acid signalling. Nature 2009, 462, 609–614. [Google Scholar]
- Moravcevic, K.; Mendrola, J.M.; Schmitz, K.R.; Wang, Y.H.; Slochower, D.; Janmey, P.A.; Lemmon, M.A. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell 2010, 143, 966–977. [Google Scholar]
- Coussens, L.; Parker, P.J.; Rhee, L.; Yang-Feng, T.L.; Chen, E.; Waterfield, M.D.; Francke, U.; Ullrich, A. Multiple, distinct forms of bovine and human protein kinase C suggest diversity in cellular signaling pathways. Science 1986, 233, 859–866. [Google Scholar]
- Parker, P.J.; Coussens, L.; Totty, N.; Rhee, L.; Young, S.; Chen, E.; Stabel, S.; Waterfield, M.D.; Ullrich, A. The complete primary structure of protein kinase C—The major phorbol ester receptor. Science 1986, 233, 853–859. [Google Scholar]
- Haeseleer, F.; Imanishi, Y.; Sokal, I.; Filipek, S.; Palczewski, K. Calcium-binding proteins: Intracellular sensors from the calmodulin superfamily. Biochem. Biophys. Res. Commun 2002, 290, 615–623. [Google Scholar]
- Meador, W.E.; Means, A.R.; Quiocho, F.A. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science 1993, 262, 1718–1721. [Google Scholar]
- Batistic, O.; Kudla, J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 2009, 1793, 985–992. [Google Scholar]
- Veeraraghavan, S.; Fagan, P.A.; Hu, H.; Lee, V.; Harper, J.F.; Huang, B.; Chazin, W.J. Structural independence of the two EF-hand domains of caltractin. J. Biol. Chem 2002, 277, 28564–28571. [Google Scholar]
- Ames, J.B.; Hendricks, K.B.; Strahl, T.; Huttner, I.G.; Hamasaki, N.; Thorner, J. Structure and calcium-binding properties of Frq1, a novel calcium sensor in the yeast Saccharomyces cerevisiae. Biochemistry 2000, 39, 12149–12161. [Google Scholar]
- Ames, J.B.; Ikura, M.; Stryer, L. Molecular structure of membrane-targeting calcium sensors in vision: Recoverin and guanylate cyclase-activating protein 2. Methods Enzymol 2000, 316, 121–132. [Google Scholar]
- Sanchez-Barrena, M.J. Estructura y Mecanismo de acción del regulador SOS3: Respuesta al estrés salino en la ruta SOS. Ph.D. Thesis, Universidad Complutense de Madrid, Madrid, Spain, 2005. [Google Scholar]
- Schapire, A.L.; Voigt, B.; Jasik, J.; Rosado, A.; Lopez-Cobollo, R.; Menzel, D.; Salinas, J.; Mancuso, S.; Valpuesta, V.; Baluska, F.; et al. Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 2008, 20, 3374–3388. [Google Scholar]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 2002, 14, S389–S400. [Google Scholar]
- Reddi, A.R.; Jensen, L.T.; Culotta, V.C. Manganese homeostasis in Saccharomyces cerevisiae. Chem. Rev 2009, 109, 4722–4732. [Google Scholar]
- Ishitani, M.; Liu, J.; Halfter, U.; Kim, C.S.; Shi, W.; Zhu, J.K. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 2000, 12, 1667–1678. [Google Scholar]
- Batistic, O.; Sorek, N.; Schultke, S.; Yalovsky, S.; Kudla, J. Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 2008, 20, 1346–1362. [Google Scholar]
- Monne, J.; Diez, Y.; Puy, J.; Galceran, J.; Nelson, A. Interpreting ion fluxes to channel arrays in monolayers. Langmuir 2007, 23, 10581–10588. [Google Scholar]
- Killian, J.A.; von Heijne, G. How proteins adapt to a membrane-water interface. Trends Biochem. Sci 2000, 25, 429–434. [Google Scholar]
- Baldwin, A.N.; Ames, J.B. Core mutations that promote the calcium-induced allosteric transition of bovine recoverin. Biochemistry 1998, 37, 17408–17419. [Google Scholar]
- Batistic, O.; Kudla, J. Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta 2012, 1820, 1283–1293. [Google Scholar]
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol 2010, 61, 593–620. [Google Scholar]
- Menendez, M.; Solis, D.; Usobiaga, P.; Laynez, J. AMP interaction sites in glycogen phosphorylase b. A thermodynamic analysis. Biophys. Chem 1985, 21, 249–260. [Google Scholar]
- Carlton, J.G.; Cullen, P.J. Coincidence detection in phosphoinositide signaling. Trends Cell Biol 2005, 15, 540–547. [Google Scholar]


© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sánchez-Barrena, M.J.; Martínez-Ripoll, M.; Albert, A. Structural Biology of a Major Signaling Network that Regulates Plant Abiotic Stress: The CBL-CIPK Mediated Pathway. Int. J. Mol. Sci. 2013, 14, 5734-5749. https://doi.org/10.3390/ijms14035734
Sánchez-Barrena MJ, Martínez-Ripoll M, Albert A. Structural Biology of a Major Signaling Network that Regulates Plant Abiotic Stress: The CBL-CIPK Mediated Pathway. International Journal of Molecular Sciences. 2013; 14(3):5734-5749. https://doi.org/10.3390/ijms14035734
Chicago/Turabian StyleSánchez-Barrena, María José, Martín Martínez-Ripoll, and Armando Albert. 2013. "Structural Biology of a Major Signaling Network that Regulates Plant Abiotic Stress: The CBL-CIPK Mediated Pathway" International Journal of Molecular Sciences 14, no. 3: 5734-5749. https://doi.org/10.3390/ijms14035734
APA StyleSánchez-Barrena, M. J., Martínez-Ripoll, M., & Albert, A. (2013). Structural Biology of a Major Signaling Network that Regulates Plant Abiotic Stress: The CBL-CIPK Mediated Pathway. International Journal of Molecular Sciences, 14(3), 5734-5749. https://doi.org/10.3390/ijms14035734