The Voltage-Dependent Anion Channel 1 (AtVDAC1) Negatively Regulates Plant Cold Responses during Germination and Seedling Development in Arabidopsis and Interacts with Calcium Sensor CBL1
Abstract
:1. Introduction
2. Results
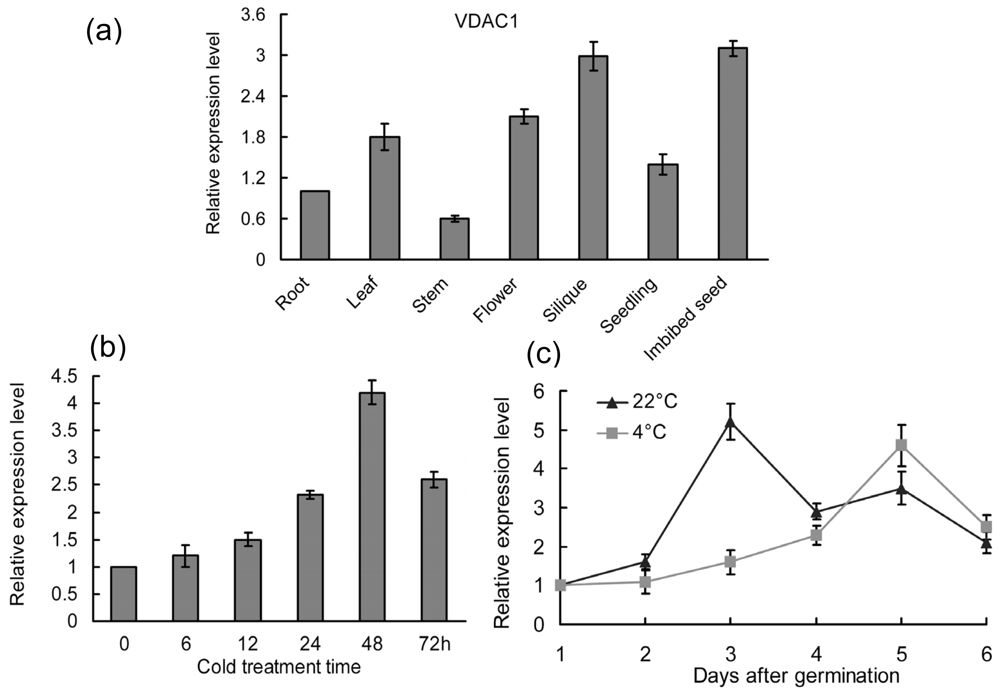
2.1. Expression Patterns of the VDAC1 in Arabidopsis
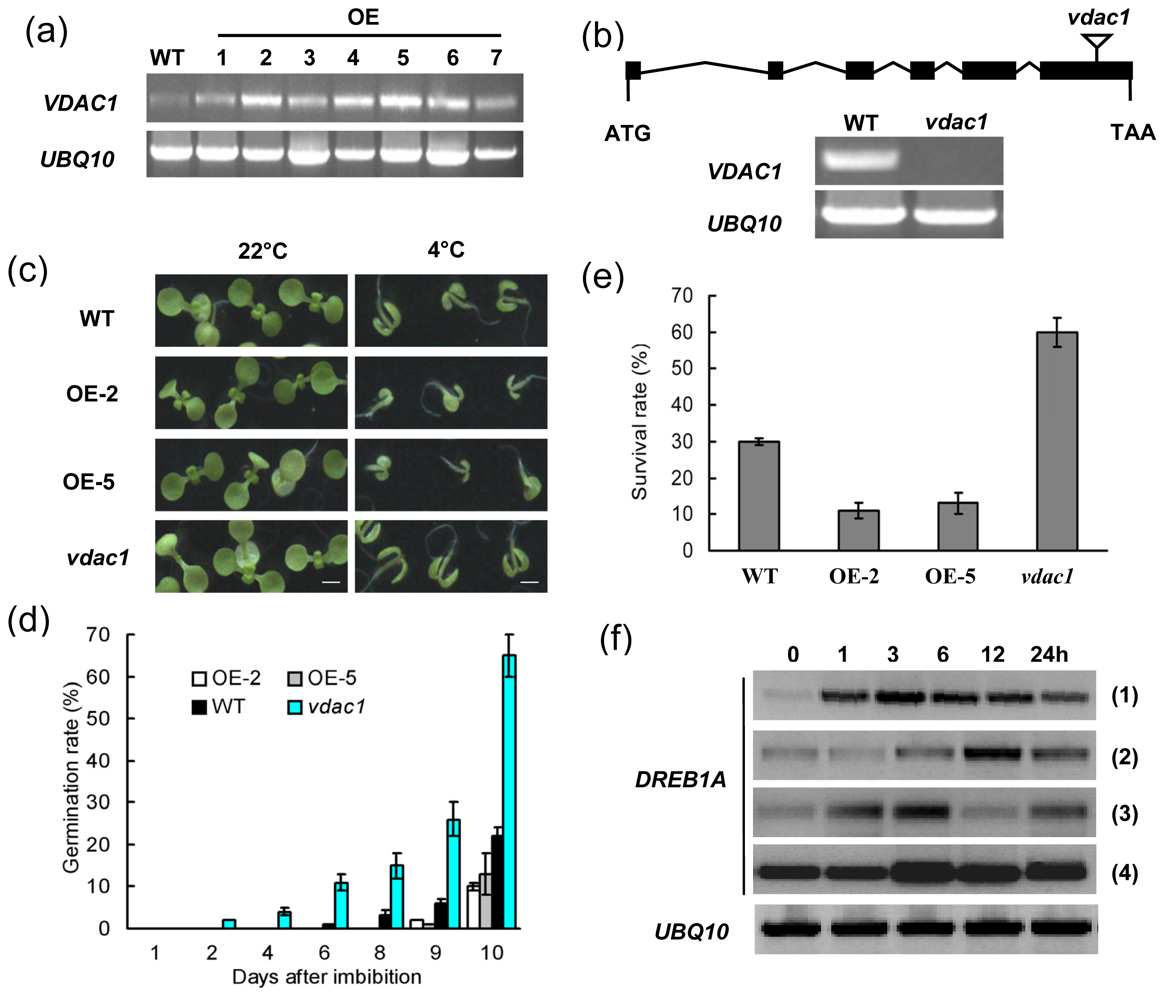
2.2. VDAC1 Overexpressing Arabidopsis Plants is Sensitive to Cold Stress
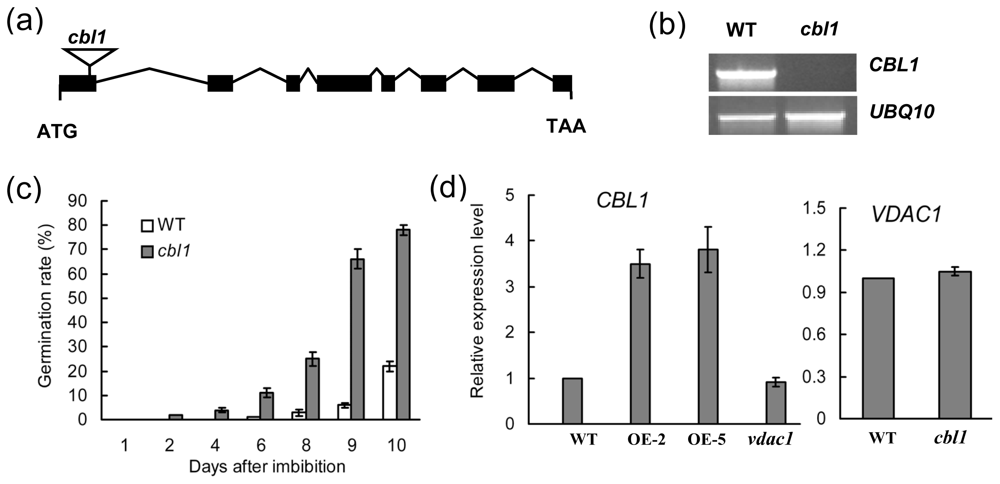
2.3. Interaction of VDAC1-CBL1
2.4. cbl1 Was Insensitive to Low Temperature during Seed Germination
3. Discussion
4. Experimental Section
4.1. Plant Material and Growth Conditions
4.2. Screening of T-DNA Insertion Mutants
4.3. RNA Isolation and Real Time PCR
4.4. Construction of Expression Vector and Isolation of Transgenic Plants
4.5. Seed Germination Assay
4.6. Yeast Two-Hybrid Screening
4.7. GST Pull-Down Assay
5. Conclusions
Acknowledgments
References
- Lee, A.C.; Xu, X.; Blachly-Dyson, E.; Forte, M.; Colombini, M. The role of yeast VDAC genes on the permeability of the mitochondrial outer membrane. J. Membr. Biol 1998, 161, 173–181. [Google Scholar]
- Sampson, M.J.; Lovell, R.S.; Craigen, W.J. The murine voltagedependent anion channel gene family. Conserved structure and function. J. Biol. Chem 1997, 272, 18966–18973. [Google Scholar]
- Clausen, C.; Ilkavets, I.; Thompson, R.; Philippar, K.; Vojta, A.; Möhlmann, T.; Neuhaus, E.; Fulgosi, H.; Soll, J. Intracellular localization of VDAC proteins in plants. Planta 2004, 220, 30–37. [Google Scholar]
- Roosens, N.; Al Bitar, F.; Jacobs, M.; Homble, F. Characterization of a cDNA encoding a rice mitochondrial voltage-dependent anion channel and its gene expression studied upon plant development and osmotic stress. BBA-Biomembranes 2000, 1463, 470–476. [Google Scholar]
- Al Bitar, F.; Roosens, N.; Smeyers, M.; Vauterin, M.; van, B.J.; Jacobs, M.; Homble, F. Sequence analysis, transcriptional and posttranscriptional regulation of the rice vdac family. BBA-Gene Struct. Expr 2003, 1625, 43–51. [Google Scholar]
- Tateda, C.; Yamashita, K.; Takahashi, F.; Kusano, T.; Takahashi, Y. Plant voltage-dependent anion channels are involved in host defense against Pseudomonas cichorii and in Bax-induced cell death. Plant Cell Rep 2009, 28, 41–51. [Google Scholar]
- Wandrey, M.; Trevaskis, B.; Brewin, N.; Udvardi, M.K. Molecular and cell biology of a family of voltage-dependent anion channel porins in Lotus japonicus. Plant Physiol 2004, 134, 182–193. [Google Scholar]
- Elkeles, A.; Breiman, A.; Zizi, M. Functional differences among wheat voltage-dependent anion channel (VDAC) isoforms expressed in yeast. Indication for the presence of a novel VDAC-modulating protein? J. Biol. Chem 1997, 272, 6252–6260. [Google Scholar]
- Wang, J.; Zhang, L.D.; Zuo, K.J.; Oian, H.M.; Cao, Y.F.; Tang, K.X. Cloning and expressional studies of the voltage-dependent anion channel gene from Brassica rapa L. J. Integr. Plant Biol 2006, 48, 197–203. [Google Scholar]
- Colombini, M. VDAC: The channel at the interface between mitochondria and the cytosol. Mol. Cell Biochem 2004, 256, 107–115. [Google Scholar]
- Homblé, F.; Krammer, E.M.; Prévost, M. Plant VDAC: Facts and speculations. Biochim. Biophys. Acta 2012, 1818, 1486–1501. [Google Scholar]
- Xu, X.; Decker, W.; Sampson, M.J.; Craigen, W.J.; Colombini, M. Mouse VDAC isoforms expressed in yeast: Channel properties and their roles in mitochondrial outer membrane permeability. J. Membr. Biol 1999, 170, 89–102. [Google Scholar]
- Shoshan-Barmatz, V.; Keinan, N.; Zaid, H. Uncovering the role of VDAC in the regulation of cell life and death. J. Bioenerg. Biomembr 2008, 40, 183–191. [Google Scholar]
- Salinas, T.; Duchêne, A.M.; Delage, L.; Nilsson, S.; Glaser, E.; Zaepfel, M.; Maréchal-Drouard, L. The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc. Natl. Acad. Sci. USA 2006, 103, 18362–18367. [Google Scholar]
- Salinas, T.; Duchêne, A.M.; Maréchal-Drouard, L. Recent advances in tRNA mitochondrial import. Trends Biochem. Sci 2008, 33, 320–329. [Google Scholar]
- Sieber, F.; Placido, A.; El Farouk-Ameqrane, S.; Duchêne, A.M.; Maréchal-Drouard, L. A protein shuttle system to target RNA into mitochondria. Nucleic Acids Res 2011, 39, e96. [Google Scholar]
- Godbole, A.; Varghese, J.; Sarin, A.; Mathew, M.K. VDAC is a conserved element of death pathways in plant and animal systems. Biochim. Biophys. Acta 2003, 1642, 87–96. [Google Scholar]
- Lee, S.M.; Hoang, M.H.; Han, H.J.; Kim, H.S.; Lee, K.; Kim, K.E.; Kim, D.H.; Lee, S.Y.; Chung, W.S. Pathogen inducible voltage-dependent anion channel (AtVDAC) isoforms are localized to mitochondria membrane in Arabidopsis. Mol. Cells 2009, 27, 321–327. [Google Scholar]
- Desai, M.K.; Mishra, R.N.; Verma, D.; Nair, S.; Sopory, S.K.; Reddy, M.K. Structural and functional analysis of a salt stress inducible gene encoding voltage dependent anion channel (VDAC) from pearl millet (Pennisetum glaucum). Plant Physiol. Biochem 2006, 44, 483–493. [Google Scholar]
- Tateda, C.; Watanabe, K.; Kusano, T.; Takahashi, Y. Molecular and genetic characterization of the gene family encoding the voltage-dependent anion channel in Arabidopsis. J. Exp. Bot 2011, 62, 4773–4785. [Google Scholar]
- Robert, N.; d’Erfurth, I.; Marmagne, A.; Erhardt, M.; Allot, M.; Boivin, K.; Gissot, L.; Monachello, D.; Michaud, M.; Duchêne, A.M.; et al. Voltage-dependent-anion-channels (VDACs) in Arabidopsis have a dual localization in the cell but show a distinct role in mitochondria. Plant Mol. Biol 2012, 78, 431–446. [Google Scholar]
- Yan, J.P.; He, H.; Tong, S.B.; Zhang, W.R.; Wang, J.M.; Li, X.F.; Yang, Y. Voltage-dependent anion channel 2 of Arabidopsis thaliana (AtVDAC2) is involved in ABA-mediated early seedling development. Int. J. Mol. Sci 2009, 10, 2476–2486. [Google Scholar]
- Cheong, Y.H.; Kim, K.N.; Pandey, G.K.; Gupta, R.; Grant, J.J.; Luan, S. CBL1, a calcium sensor that differentially regulates salt, drought and cold responses in Arabidopsis. Plant Cell 2003, 15, 1833–1845. [Google Scholar]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 2004, 136, 2621–2632. [Google Scholar]
- Kmita, H.; Budzińska, M.; Stobienia, O. Modulation of the voltagedependent anion-selective channel by cytoplasmic proteins from wild type and the channel depleted cells of Saccharomyces cerevisiae. Acta Biochim. Polonica 2003, 50, 415–424. [Google Scholar]
- Abu-Hamad, S.; Sivan, S.; Shoshan-Barmatz, V. The expression level of the voltage-dependent anion channel controls life and death of the cell. Proc. Natl. Acad. Sci. USA 2006, 103, 5787–5792. [Google Scholar]
- Krauskopf, A.; Eriksson, O.; Craigen, W.J.; Forte, M.A.; Bernardi, P. Properties of the permeability transition in VDAC1(−/−) mitochondria. Biochim. Biophys. Acta 2006, 1757, 590–595. [Google Scholar]
- Perl, M. ATP synthesis and utilization in the early stage of seed germination in relation to seed dormancy and quality. Physiol. Plantarum 1986, 66, 177–182. [Google Scholar]
- Shoshan-Barmatz, V.; Israelson, A. The voltage-dependent anion channel in endoplasmic/sarcoplasmic reticulum: Characterization, modulation and possible function. J. Membr. Biol 2005, 204, 57–66. [Google Scholar]
- Balasubramanian, R.; Karve, A.; Kandasamy, M.; Meagher, R.B.; Moore, B. A role for F-actin in hexokinase-mediated glucose signaling. Plant Physiol 2007, 145, 1423–1434. [Google Scholar]
- Gincel, D.; Zaid, H.; Shoshan-Barmatz, V. Calcium binding and translocation by the voltage dependent anion channel: A possible regulatory mechanism in mitochondrial function. Biochem. J 2001, 358, 147–155. [Google Scholar]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 2002, 14, S389–S400. [Google Scholar]
- Trewavas, A.J.; Malho, R. Ca2+ signaling in plant cells: The big network! Curr. Opin. Plant Biol 1998, 1, 428–433. [Google Scholar]
- Swidzinski, J.A.; Sweetlove, L.J.; Leaver, C.J. A custom microarray analysis of gene expression during programmed cell death in Arabidopsis thaliana. Plant J 2002, 30, 431–446. [Google Scholar]
- Swidzinski, J.A.; Leaver, C.J.; Sweetlove, L.J. A proteomic analysis of plant programmed cell death. Phytochemistry 2004, 65, 1829–1838. [Google Scholar]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 1980, 8, 4321–4325. [Google Scholar]
- Li, Z.Y.; Xu, Z.S.; He, G.Y.; Yang, G.X.; Chen, M.; Li, L.C.; Ma, Y.Z. A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. Biophys. Res. Commun 2012, 426, 522–527. [Google Scholar]
- Zhang, H.Y.; Mao, X.G.; Jing, R.L.; Chang, X.P.; Xie, H. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. J. Exp. Bot 2011, 62, 975–988. [Google Scholar]
- Xu, Z.S.; Xia, L.Q.; Chen, M.; Cheng, X.G.; Zhang, R.Y.; Li, L.C.; Zhao, Y.X.; Lu, Y.; Ni, Z.Y.; Liu, L.; et al. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol 2007, 65, 719–732. [Google Scholar]

© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, Z.-Y.; Xu, Z.-S.; He, G.-Y.; Yang, G.-X.; Chen, M.; Li, L.-C.; Ma, Y. The Voltage-Dependent Anion Channel 1 (AtVDAC1) Negatively Regulates Plant Cold Responses during Germination and Seedling Development in Arabidopsis and Interacts with Calcium Sensor CBL1. Int. J. Mol. Sci. 2013, 14, 701-713. https://doi.org/10.3390/ijms14010701
Li Z-Y, Xu Z-S, He G-Y, Yang G-X, Chen M, Li L-C, Ma Y. The Voltage-Dependent Anion Channel 1 (AtVDAC1) Negatively Regulates Plant Cold Responses during Germination and Seedling Development in Arabidopsis and Interacts with Calcium Sensor CBL1. International Journal of Molecular Sciences. 2013; 14(1):701-713. https://doi.org/10.3390/ijms14010701
Chicago/Turabian StyleLi, Zhi-Yong, Zhao-Shi Xu, Guang-Yuan He, Guang-Xiao Yang, Ming Chen, Lian-Cheng Li, and Youzhi Ma. 2013. "The Voltage-Dependent Anion Channel 1 (AtVDAC1) Negatively Regulates Plant Cold Responses during Germination and Seedling Development in Arabidopsis and Interacts with Calcium Sensor CBL1" International Journal of Molecular Sciences 14, no. 1: 701-713. https://doi.org/10.3390/ijms14010701



