Function and 3D Structure of the N-Glycans on Glycoproteins
Abstract
:1. Introduction
2. Conserved Glycan Mediates Inter-Subunit Interactions of Proteins (Fc Fragment and Influenza Neuraminidase)
2.1. N-Glycan on Fc Fragment Affects Intra- and Inter-Molecular Interactions
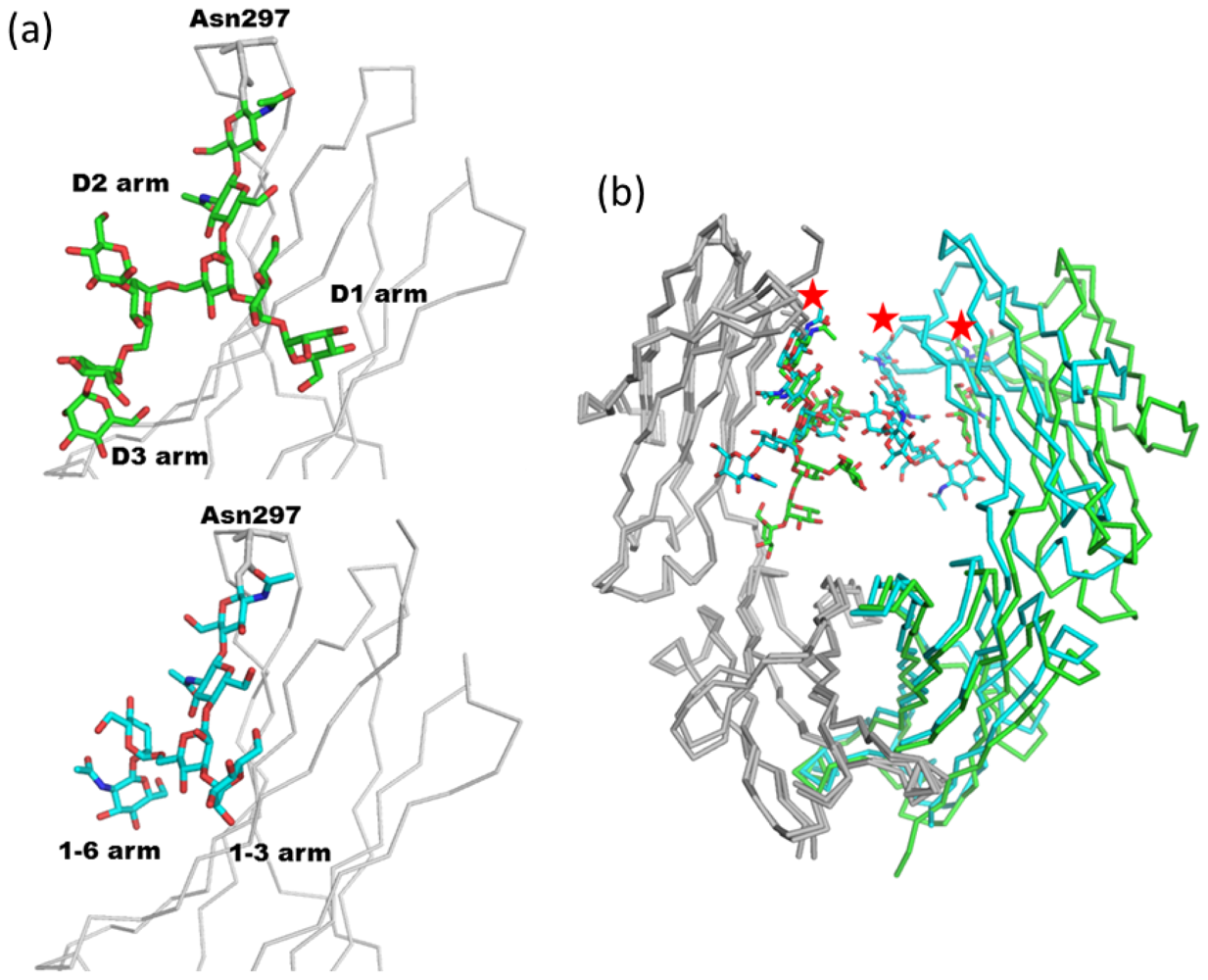
2.1.1. Overview of N-Glycan of Fc Fragment
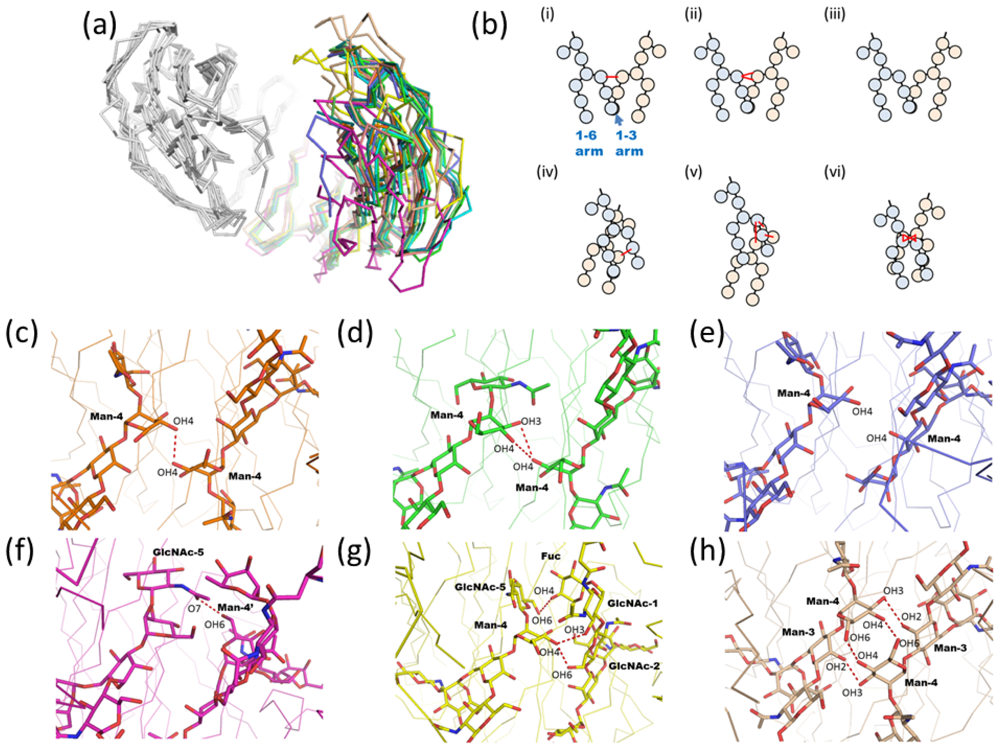
2.1.2. Glycoform Affects the Relative Interdomain Angles of the Fc Fragment
2.1.3. Intra-molecular Carbohydrate-Carbohydrate Interaction
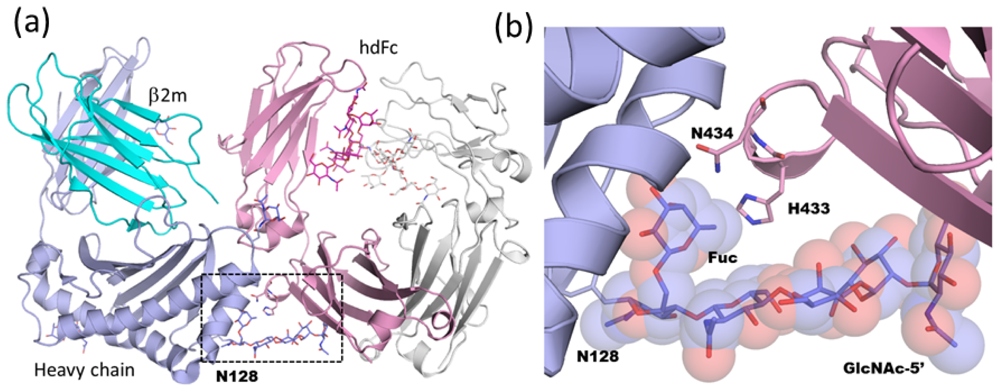
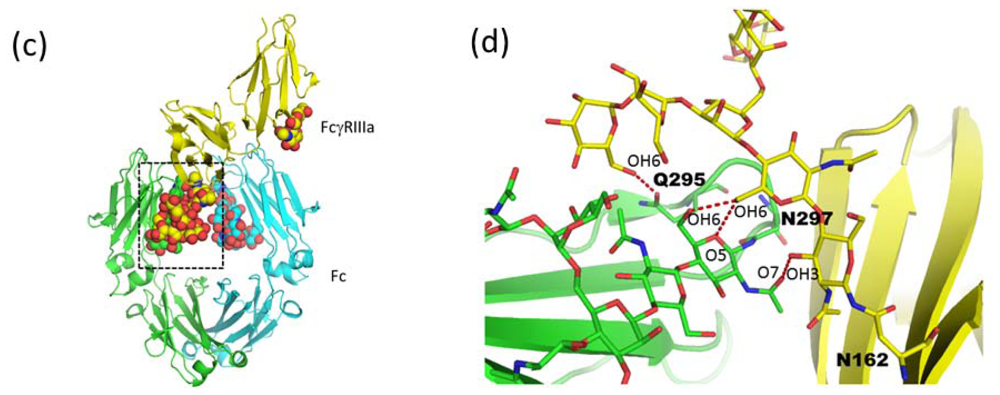
2.1.4. Carbohydrate-Assisted Intermolecular Interaction (Neonatal Fc Receptor and Fcγ Receptor IIIa)
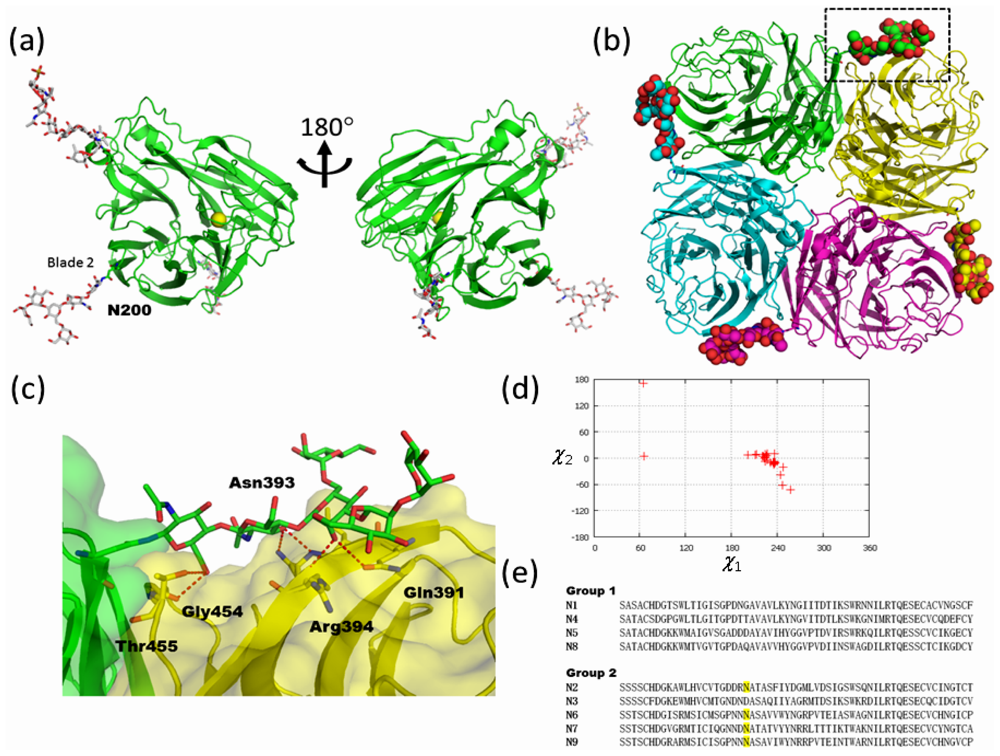
2.2. High-Mannose Type Glycan on Group 2 Influenza Virus Neuraminidase
3. Immature High-Mannose Type Glycans Contribute to Inter-Subunit and Inter-Domain Interactions
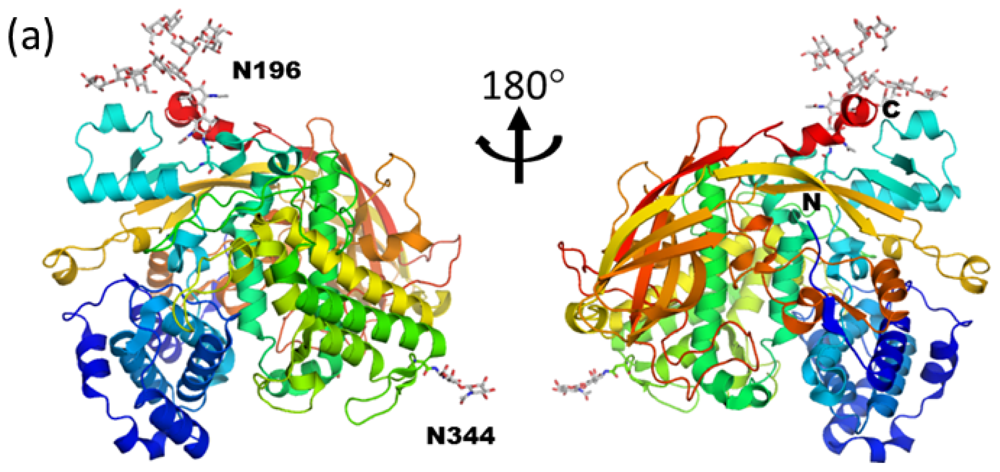
3.1. Monoglucosylated High-Mannose Type Glycan Stabilizes Hexamer Formation of Arylphorin from Antheraea pernyi
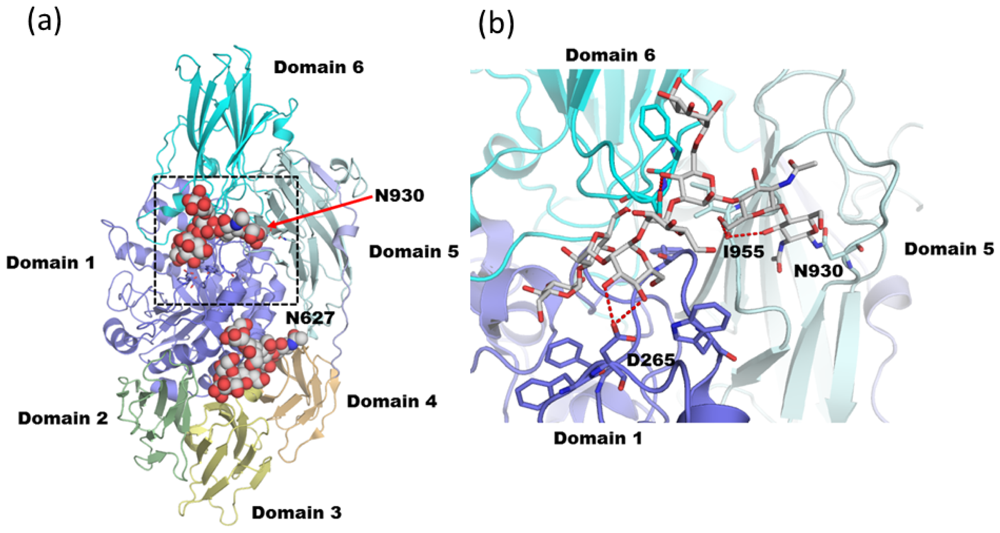
3.2. Diglucosylated N-Glycan Stabilizes Inter-Domain Interaction of β-Dalactosidase from Trichoderma reesei
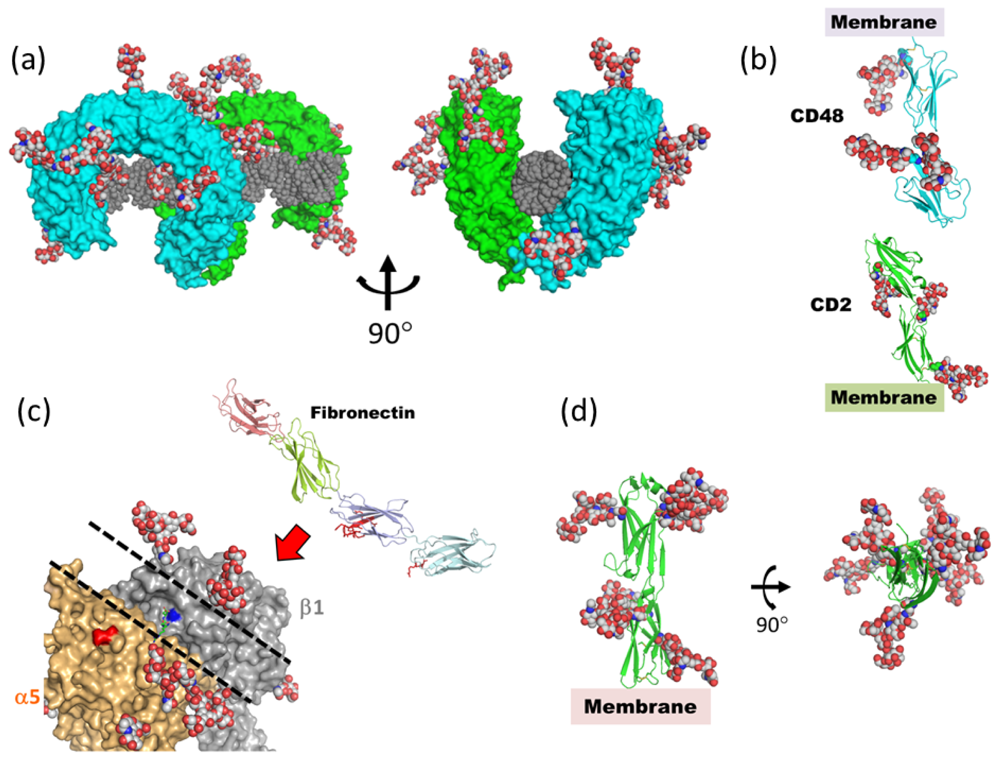
4. What Is the Function of Mobile/Disordered N-Glycans?
5. Future Perspective
Abbreviations
| GlcNAc | N-acetyl-d-glucosamine |
| Man | d-mannose |
| Gal | d-galactose |
| Sia | sialic acid |
| Glc | d-glucose |
| Fuc | l-fucose |
| GnT | N-acetylglucosaminyltransferase |
| CHO | Chinese hamster ovary |
| HEK | human embryonic kidney |
| IgG | immunoglobulin G |
| Fc | crystallizable fragment |
| ADCC | antibody-dependent cellular cytotoxicity |
| FcRn | neonatal Fc receptor |
| NA | neuraminidase |
| APA | Antheraea pernyi arylphorin |
| Tr-β-gal | Trichoderma reesei β-galactosidase |
| TLR | Toll-like receptor |
| ICAM | intracellular adhesion molecule |
References
- Helenius, A.; Aebi, M. Intracellular functions of N-linked glycans. Science 2001, 291, 2364–2369. [Google Scholar]
- Apweiler, R.; Hermjakob, H.; Sharon, N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1999, 1473, 4–8. [Google Scholar]
- Nishikawa, A.; Ihara, Y.; Hatakeyama, M.; Kangawa, K.; Taniguchi, N. Purification, cDNA cloning, and expression of UDP-N-acetylglucosamine:β-d-mannoside β-1,4-Nacetylglucosaminyltransferase III from rat kidney. J. Biol. Chem 1992, 267, 18199–18204. [Google Scholar]
- Cummings, R.D.; Trowbridge, I.S.; Kornfeld, S. A mouse lymphoma cell line resistant to the leukoagglutinating lectin from Phaseolus vulgaris is deficient in UDP-GlcNAc:α-d-mannoside- β-1,6-N-acetylglucosaminyltransferase. J. Biol. Chem 1982, 257, 13421–13427. [Google Scholar]
- Uozumi, N.; Yanagidani, S.; Miyoshi, E.; Ihara, Y.; Sakuma, T.; Gao, C.X.; Teshima, T.; Fujii, S.; Shiba, T.; Taniguchi, N. Purification and cDNA cloning of porcine brain GDP-l-Fuc:Nacetyl- β-d-glucosaminide α1→6 fucosyltransferase. J. Biol. Chem 1996, 271, 27810–27817. [Google Scholar]
- Wyss, D.F.; Wagner, G. The structural role of sugars in glycoproteins. Curr. Opin. Biotechnol 1996, 7, 409–416. [Google Scholar]
- Kwong, P.D.; Wyatt, R.; Desjardins, E.; Robinson, J.; Culp, J.S.; Hellmig, B.D.; Sweet, R.W.; Sodroski, J.; Hendrickson, W.A. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J. Biol. Chem 1999, 274, 4115–4123. [Google Scholar]
- Nakata, Z.; Nagae, M.; Yasui, N.; Bujo, H.; Nogi, T.; Takagi, J. Crystallization and preliminary crystallographic analysis of human LR11 Vps10p domain. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun 2011, 67, 129–132. [Google Scholar]
- Petrescu, A.J.; Milac, A.L.; Petrescu, S.M.; Dwek, R.A.; Wormald, M.R. Statistical analysis of the protein environment of N-glycosylation sites: Implications for occupancy, structure, and folding. Glycobiology 2004, 14, 103–114. [Google Scholar]
- Stanley, P. Chinese hamster ovary cell mutants with multiple glycosylation defects for production of glycoproteins with minimal carbohydrate heterogeneity. Mol. Cell. Biol 1989, 9, 377–383. [Google Scholar]
- Reeves, P.J.; Callewaert, N.; Contreras, R.; Khorana, H.G. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. USA 2002, 99, 13419–13424. [Google Scholar]
- Chang, V.T.; Crispin, M.; Aricescu, A.R.; Harvey, D.J.; Nettleship, J.E.; Fennelly, J.A.; Yu, C.; Boles, K.S.; Evans, E.J.; Stuart, D.I.; et al. Glycoprotein structural genomics: Solving the glycosylation problem. Structure 2007, 15, 267–273. [Google Scholar]
- Petrescu, A.J.; Petrescu, S.M.; Dwek, R.A.; Wormald, M.R. A statistical analysis of N- and O-glycan linkage conformations from crystallographic data. Glycobiology 1999, 9, 343–352. [Google Scholar]
- Wormald, M.R.; Petrescu, A.J.; Pao, Y.L.; Glithero, A.; Elliott, T.; Dwek, R.A. Conformational studies of oligosaccharides and glycopeptides: Complementarity of NMR, X-ray crystallography, and molecular modelling. Chem. Rev 2002, 102, 371–386. [Google Scholar]
- Lütteke, T.; Bohne-Lang, A.; Loss, A.; Goetz, T.; Frank, M.; von der Lieth, C.W. GLYCOSCIENCES.de: An Internet portal to support glycomics and glycobiology research. Glycobiology 2006, 16, 71R–81R. [Google Scholar]
- Nakahara, T.; Hashimoto, R.; Nakagawa, H.; Monde, K.; Miura, N.; Nishimura, S. Glycoconjugate Data Bank: Structures—An annotated glycan structure database and N-glycan primary structure verification service. Nucleic Acids Res 2008, 36, D368–371. [Google Scholar]
- Deisenhofer, J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-Å resolution. Biochemistry 1981, 20, 2361–2370. [Google Scholar]
- Martin, W.L.; West, A.P., Jr; Gan, L.; Bjorkman, P.J. Crystal structure at 2.8 Å of an FcRn/heterodimeric Fc complex: Mechanism of pH-dependent binding. Mol. Cell 2001, 7, 867–877. [Google Scholar]
- Harris, L.J.; Larson, S.B.; Hasel, K.W.; McPherson, A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 1997, 36, 1581–1597. [Google Scholar]
- Saphire, E.O.; Stanfield, R.L.; Crispin, M.D.; Parren, P.W.; Rudd, P.M.; Dwek, R.A.; Burton, D.R.; Wilson, I.A. Contrasting IgG structures reveal extreme asymmetry and flexibility. J. Mol. Biol 2002, 319, 9–18. [Google Scholar]
- Dwek, R.A.; Lellouch, A.C.; Wormald, M.R. Glycobiology: “The function of sugar in the IgG molecule”. J. Anat 1995, 187 Pt 2, 279–292. [Google Scholar]
- Shields, R.L.; Lai, J.; Keck, R.; O’Connell, L.Y.; Hong, K.; Meng, Y.G.; Weikert, S.H.; Presta, L.G. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcγ RIII and antibody-dependent cellular toxicity. J. Biol. Chem 2002, 277, 26733–26740. [Google Scholar]
- Nimmerjahn, F.; Ravetch, J.V. Divergent immunoglobulin G subclass activity through selective Fc receptor binding. Science 2005, 310, 1510–1512. [Google Scholar]
- Parekh, R.B.; Dwek, R.A.; Sutton, B.J.; Fernandes, D.L.; Leung, A.; Stanworth, D.; Rademacher, T.W.; Mizuochi, T.; Taniguchi, T.; Matsuta, K.; et al. Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature 1985, 316, 452–457. [Google Scholar]
- Samuelsson, A.; Towers, T.L.; Ravetch, J.V. Anti-inflammatory activity of IVIG mediated through the inhibitory Fc receptor. Science 2001, 291, 484–486. [Google Scholar]
- Umaña, P.; Jean-Mairet, J.; Moudry, R.; Amstutz, H.; Bailey, J.E. Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat. Biotechnol 1999, 17, 176–180. [Google Scholar]
- Shinkawa, T.; Nakamura, K.; Yamane, N.; Shoji-Hosaka, E.; Kanda, Y.; Sakurada, M.; Uchida, K.; Anazawa, H.; Satoh, M.; Yamasaki, M.; et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem 2003, 278, 3466–3473. [Google Scholar]
- Sutton, B.J.; Phillips, D.C. The three-dimensional structure of the carbohydrate within the Fc fragment of immunoglobulin G. Biochem. Soc. Trans 1983, 11, 130–132. [Google Scholar]
- Matsumiya, S.; Yamaguchi, Y.; Saito, J.; Nagano, M.; Sasakawa, H.; Otaki, S.; Satoh, M.; Shitara, K.; Kato, K. Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J. Mol. Biol 2007, 368, 767–779. [Google Scholar]
- Crispin, M.; Bowden, T.A.; Coles, C.H.; Harlos, K.; Aricescu, A.R.; Harvey, D.J.; Stuart, D.I.; Jones, E.Y. Carbohydrate and domain architecture of an immature antibody glycoform exhibiting enhanced effector functions. J. Mol. Biol 2009, 387, 1061–1066. [Google Scholar]
- Krapp, S.; Mimura, Y.; Jefferis, R.; Huber, R.; Sondermann, P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J. Mol. Biol 2003, 325, 979–989. [Google Scholar]
- Mimura, Y.; Sondermann, P.; Ghirlando, R.; Lund, J.; Young, S.P.; Goodall, M.; Jefferis, R. Role of oligosaccharide residues of IgG1-Fc in FcγRIIb binding. J. Biol. Chem 2001, 276, 45539–45547. [Google Scholar]
- Oganesyan, V.; Damschroder, M.M.; Woods, R.M.; Cook, K.E.; Wu, H.; Dall’acqua, W.F. Structural characterization of a human Fc fragment engineered for extended serum half-life. Mol. Immunol 2009, 46, 1750–1755. [Google Scholar]
- Oganesyan, V.; Gao, C.; Shirinian, L.; Wu, H.; Dall’Acqua, W.F. Structural characterization of a human Fc fragment engineered for lack of effector functions. Acta Crystallogr. D Biol. Crystallogr 2008, 64, 700–704. [Google Scholar]
- Oganesyan, V.; Damschroder, M.M.; Leach, W.; Wu, H.; Dall’Acqua, W.F. Structural characterization of a mutated, ADCC-enhanced human Fc fragment. Mol. Immunol 2008, 45, 1872–1882. [Google Scholar]
- DeLano, W.L.; Ultsch, M.H.; de Vos, A.M.; Wells, J.A. Convergent solutions to binding at a protein-protein interface. Science 2000, 287, 1279–1283. [Google Scholar]
- Idusogie, E.E.; Presta, L.G.; Gazzano-Santoro, H.; Totpal, K.; Wong, P.Y.; Ultsch, M.; Meng, Y.G.; Mulkerrin, M.G. Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc. J. Immunol 2000, 164, 4178–4184. [Google Scholar]
- Branden, B.C. Department of Natural Sciences, Bowie State University: Bowie, MD, USA; Unpublished Work; 2009. [Google Scholar]
- Saphire, E.O.; Parren, P.W.; Pantophlet, R.; Zwick, M.B.; Morris, G.M.; Rudd, P.M.; Dwek, R.A.; Stanfield, R.L.; Burton, D.R.; Wilson, I.A. Crystal structure of a neutralizing human IgG against HIV-1: A template for vaccine design. Science 2001, 293, 1155–1159. [Google Scholar]
- Moiani, D.; Salvalaglio, M.; Cavallotti, C.; Bujacz, A.; Redzynia, I.; Bujacz, G.; Dinon, F.; Pengo, P.; Fassina, G. Structural characterization of a Protein A mimetic peptide dendrimer bound to human IgG. J. Phys. Chem. B 2009, 113, 16268–16275. [Google Scholar]
- Kolenko, P.; Dohnálek, J.; Dušková, J.; Skálová, T.; Collard, R.; Hašek, J. New insights into intra- and intermolecular interactions of immunoglobulins: Crystal structure of mouse IgG2b-Fc at 2.1-Å resolution. Immunology 2009, 126, 378–385. [Google Scholar]
- Girardi, E.; Holdom, M.D.; Davies, A.M.; Sutton, B.J.; Beavil, A.J. The crystal structure of rabbit IgG-Fc. Biochem. J 2009, 417, 77–83. [Google Scholar]
- Raju, T.S.; Mulkerrin, M.G.; Parker, M.; de Vos, A.M.; Gazzano-Santoro, H.; Totpal, K.; Ultsch, M.H. Department of Protein Engineering, Genentech Inc: South San Francisco, CA, USA; Unpublished Work; 2003. [Google Scholar]
- Radaev, S.; Motyka, S.; Fridman, W.H.; Sautes-Fridman, C.; Sun, P.D. The structure of a human type III Fcγ receptor in complex with Fc. J. Biol. Chem 2001, 276, 16469–16477. [Google Scholar]
- Sondermann, P.; Huber, R.; Oosthuizen, V.; Jacob, U. The 3.2-Å crystal structure of the human IgG1 Fc fragment-FcγRIII complex. Nature 2000, 406, 267–273. [Google Scholar]
- Ferrara, C.; Grau, S.; Jäger, C.; Sondermann, P.; Brünker, P.; Waldhauer, I.; Hennig, M.; Ruf, A.; Rufer, A.C.; Stihle, M.; et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc. Natl. Acad. Sci. USA 2011, 108, 12669–12674. [Google Scholar]
- Mizushima, T.; Yagi, H.; Takemoto, E.; Shibata-Koyama, M.; Isoda, Y.; Iida, S.; Masuda, K.; Satoh, M.; Kato, K. Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans. Genes Cells 2011, 16, 1071–1080. [Google Scholar]
- Lütteke, T. Analysis and validation of carbohydrate three-dimensional structures. Acta Crystallogr. D Biol. Crystallogr 2009, 65, 156–168. [Google Scholar]
- Wormald, M.R.; Rudd, P.M.; Harvey, D.J.; Chang, S.C.; Scragg, I.G.; Dwek, R.A. Variations in oligosaccharide-protein interactions in immunoglobulin G determine the site-specific glycosylation profiles and modulate the dynamic motion of the Fc oligosaccharides. Biochemistry 1997, 36, 1370–1380. [Google Scholar]
- Yamaguchi, Y.; Kato, K.; Shindo, M.; Aoki, S.; Furusho, K.; Koga, K.; Takahashi, N.; Arata, Y.; Shimada, I. Dynamics of the carbohydrate chains attached to the Fc portion of immunoglobulin G as studied by NMR spectroscopy assisted by selective 13C labeling of the glycans. J. Biomol. NMR 1998, 12, 385–394. [Google Scholar]
- Barb, A.W.; Prestegard, J.H. NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat. Chem. Biol 2011, 7, 147–153. [Google Scholar]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., III; de Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Cα geometry: φ, ϕ and Cβ deviation. Proteins 2003, 50, 437–450. [Google Scholar]
- Kolenko, P.; Skálová, T.; Dohnálek, J.; Hašek, J. L-fucose in crystal structures of IgG-Fc: Reinterpretation of experimental data. Collect. Czechoslov. Chem. Commun 2008, 73, 608–615. [Google Scholar]
- Yamaguchi, Y.; Nishimura, M.; Nagano, M.; Yagi, H.; Sasakawa, H.; Uchida, K.; Shitara, K.; Kato, K. Glycoform-dependent conformational alteration of the Fc region of human immunoglobulin G1 as revealed by NMR spectroscopy. Biochim. Biophys. Acta 2006, 1760, 693–700. [Google Scholar]
- Kanda, Y.; Yamada, T.; Mori, K.; Okazaki, A.; Inoue, M.; Kitajima-Miyama, K.; Kuni-Kamochi, R.; Nakano, R.; Yano, K.; Kakita, S.; et al. Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: The high-mannose, hybrid, and complex types. Glycobiology 2007, 17, 104–118. [Google Scholar]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr 2004, 60, 2256–2268. [Google Scholar]
- Burmeister, W.P.; Huber, A.H.; Bjorkman, P.J. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature 1994, 372, 379–383. [Google Scholar]
- Martin, W.L.; Bjorkman, P.J. Characterization of the 2:1 complex between the class I MHC-related Fc receptor and its Fc ligand in solution. Biochemistry 1999, 38, 12639–12647. [Google Scholar]
- Sánchez, L.M.; Penny, D.M.; Bjorkman, P.J. Stoichiometry of the interaction between the major histocompatibility complex-related Fc receptor and its Fc ligand. Biochemistry 1999, 38, 9471–9476. [Google Scholar]
- Clynes, R.A.; Towers, T.L.; Presta, L.G.; Ravetch, J.V. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat. Med 2000, 6, 443–446. [Google Scholar]
- Cartron, G.; Dacheux, L.; Salles, G.; Solal-Celigny, P.; Bardos, P.; Colombat, P.; Watier, H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcγRIIIa gene. Blood 2002, 99, 754–758. [Google Scholar]
- Liu, C.; Eichelberger, M.C.; Compans, R.W.; Air, G.M. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J. Virol 1995, 69, 1099–1106. [Google Scholar]
- A revision of the system of nomenclature for influenza viruses: A WHO memorandum. Bull. World Health Organ 1980, 58, 585–591.
- Russell, R.J.; Haire, L.F.; Stevens, D.J.; Collins, P.J.; Lin, Y.P.; Blackburn, G.M.; Hay, A.J.; Gamblin, S.J.; Skehel, J.J. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 2006, 443, 45–49. [Google Scholar]
- Blok, J.; Air, G.M.; Laver, W.G.; Ward, C.W.; Lilley, G.G.; Woods, E.F.; Roxburgh, C.M.; Inglis, A.S. Studies on the size, chemical composition, and partial sequence of the neuraminidase (NA) from type A influenza viruses show that the N-terminal region of the NA is not processed and serves to anchor the NA in the viral membrane. Virology 1982, 119, 109–121. [Google Scholar]
- Air, G.M.; Laver, W.G. The neuraminidase of influenza virus. Proteins 1989, 6, 341–356. [Google Scholar]
- Murti, K.G.; Webster, R.G. Distribution of hemagglutinin and neuraminidase on influenza virions as revealed by immunoelectron microscopy. Virology 1986, 149, 36–43. [Google Scholar]
- Von Itzstein, M.; Wu, W.Y.; Kok, G.B.; Pegg, M.S.; Dyason, J.C.; Jin, B.; van Phan, T.; Smythe, M.L.; White, H.F.; Oliver, S.W.; et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 1993, 363, 418–423. [Google Scholar]
- Kim, C.U.; Lew, W.; Williams, M.A.; Liu, H.; Zhang, L.; Swaminathan, S.; Bischofberger, N.; Chen, M.S.; Mendel, D.B.; Tai, C.Y.; et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: Design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J. Am. Chem. Soc 1997, 119, 681–690. [Google Scholar]
- Varghese, J.N.; Laver, W.G.; Colman, P.M. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature 1983, 303, 35–40. [Google Scholar]
- Baker, A.T.; Varghese, J.N.; Laver, W.G.; Air, G.M.; Colman, P.M. Three-dimensional structure of neuraminidase of subtype N9 from an avian influenza virus. Proteins 1987, 2, 111–117. [Google Scholar]
- Lee, B.; Richards, F.M. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol 1971, 55, 379–400. [Google Scholar]
- Burmeister, W.P.; Ruigrok, R.W.; Cusack, S. The 2.2 Å resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J 1992, 11, 49–56. [Google Scholar]
- Varghese, J.N.; Colman, P.M. Three-dimensional structure of the neuraminidase of influenza virus A/Tokyo/3/67 at 2.2 Å resolution. J. Mol. Biol 1991, 221, 473–486. [Google Scholar]
- White, C.L.; Janakiraman, M.N.; Laver, W.G.; Philippon, C.; Vasella, A.; Air, G.M.; Luo, M. A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies. J. Mol. Biol 1995, 245, 623–634. [Google Scholar]
- Rudino-Pinera, E.; Tunnah, P.; Crennell, S.J.; Webster, R.G.; Laver, W.G.; Garman, E.F. Department of Biochemistry, University of Oxford: South Parks Road, Oxford, UK; Unpublished Work; 2006. [Google Scholar]
- Smith, B.J.; Colman, P.M.; von Itzstein, M.; Danylec, B.; Varghese, J.N. Analysis of inhibitor binding in influenza virus neuraminidase. Protein Sci 2001, 10, 689–696. [Google Scholar]
- Malby, R.L.; McCoy, A.J.; Kortt, A.A.; Hudson, P.J.; Colman, P.M. Three-dimensional structures of single-chain Fv-neuraminidase complexes. J. Mol. Biol 1998, 279, 901–910. [Google Scholar]
- Smith, B.J.; Huyton, T.; Joosten, R.P.; McKimm-Breschkin, J.L.; Zhang, J.G.; Luo, C.S.; Lou, M.Z.; Labrou, N.E.; Garrett, T.P. Structure of a calcium-deficient form of influenza virus neuraminidase: Implications for substrate binding. Acta Crystallogr. D Biol. Crystallogr 2006, 62, 947–952. [Google Scholar]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. Essentials of Glycobiology, 2nd ed; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Ryu, K.S.; Lee, J.O.; Kwon, T.H.; Choi, H.H.; Park, H.S.; Hwang, S.K.; Lee, Z.W.; Lee, K.B.; Han, Y.H.; Choi, Y.S.; et al. The presence of monoglucosylated N196-glycan is important for the structural stability of storage protein, arylphorin. Biochem. J 2009, 421, 87–96. [Google Scholar]
- Burmester, T.; Scheller, K. Common origin of arthropod tyrosinase, arthropod hemocyanin, insect hexamerin, and dipteran arylphorin receptor. J. Mol. Evol 1996, 42, 713–728. [Google Scholar]
- Telfer, W.H.; Kunkel, J.G. The function and evolution of insect storage hexamers. Annu. Rev. Entomol 1991, 36, 205–228. [Google Scholar]
- Jung, H.I.; Kim, Y.H.; Kim, S. Structural basis for the presence of a monoglucosylated oligosaccharide in mature glycoproteins. Biochem. Biophys. Res. Commun 2005, 331, 100–106. [Google Scholar]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res 2009, 37, D233–238. [Google Scholar]
- Seiboth, B.; Hartl, L.; Salovuori, N.; Lanthaler, K.; Robson, G.D.; Vehmaanperä, J.; Penttilä, M.E.; Kubicek, C.P. Role of the bga1-encoded extracellular β-galactosidase of Hypocrea jecorina in cellulase induction by lactose. Appl. Environ. Microbiol 2005, 71, 851–857. [Google Scholar]
- Gamauf, C.; Marchetti, M.; Kallio, J.; Puranen, T.; Vehmaanperä, J.; Allmaier, G.; Kubicek, C.P.; Seiboth, B. Characterization of the bga1-encoded glycoside hydrolase family 35 β-galactosidase of Hypocrea jecorina with galacto-β-D-galactanase activity. FEBS J 2007, 274, 1691–1700. [Google Scholar]
- Maksimainen, M.; Hakulinen, N.; Kallio, J.M.; Timoharju, T.; Turunen, O.; Rouvinen, J. Crystal structures of Trichoderma reesei β-galactosidase reveal conformational changes in the active site. J. Struct. Biol 2011, 174, 156–163. [Google Scholar]
- Maksimainen, M.; Timoharju, T.; Kallio, J.M.; Hakulinen, N.; Turunen, O.; Rouvinen, J. Crystallization and preliminary diffraction analysis of a β-galactosidase from Trichoderma reesei. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun 2009, 65, 767–769. [Google Scholar]
- Hakomori, S. Glycosylation defining cancer malignancy: New wine in an old bottle. Proc. Natl. Acad. Sci. USA 2002, 99, 10231–10233. [Google Scholar]
- Infectious Disease Movement in a Borderless World: Workshop Summary, 2010/10/15 ed; National Academies Press (US): Washington, DC, USA, 2010.
- Janeway, C.A., Jr; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol 2002, 20, 197–216. [Google Scholar]
- Beutler, B. Innate immunity: An overview. Mol. Immunol 2004, 40, 845–859. [Google Scholar]
- Takeda, K.; Akira, S. Toll-like receptors in innate immunity. Int. Immunol 2005, 17, 1–14. [Google Scholar]
- Choe, J.; Kelker, M.S.; Wilson, I.A. Crystal structure of human Toll-like receptor 3 (TLR3) ectodomain. Science 2005, 309, 581–585. [Google Scholar]
- Liu, L.; Botos, I.; Wang, Y.; Leonard, J.N.; Shiloach, J.; Segal, D.M.; Davies, D.R. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 2008, 320, 379–381. [Google Scholar]
- Rudd, P.M.; Wormald, M.R.; Stanfield, R.L.; Huang, M.; Mattsson, N.; Speir, J.A.; DiGennaro, J.A.; Fetrow, J.S.; Dwek, R.A.; Wilson, I.A. Roles for glycosylation of cell surface receptors involved in cellular immune recognition. J. Mol. Biol 1999, 293, 351–366. [Google Scholar]
- Bodian, D.L.; Jones, E.Y.; Harlos, K.; Stuart, D.I.; Davis, S.J. Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 Å resolution. Structure 1994, 2, 755–766. [Google Scholar]
- Evans, E.J.; Castro, M.A.; O’Brien, R.; Kearney, A.; Walsh, H.; Sparks, L.M.; Tucknott, M.G.; Davies, E.A.; Carmo, A.M.; van der Merwe, P.A.; et al. Crystal structure and binding properties of the CD2 and CD244 (2B4)-binding protein, CD48. J. Biol. Chem 2006, 281, 29309–29320. [Google Scholar]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar]
- Nagae, M.; Re, S.; Mihara, E.; Nogi, T.; Sugita, Y.; Takagi, J. Crystal structure of α5β1 integrin ectodomain: Atomic details of the fibronectin receptor. J. Cell. Biol 2012, 197, 131–140. [Google Scholar]
- Leahy, D.J.; Aukhil, I.; Erickson, H.P. 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 1996, 84, 155–164. [Google Scholar]
- Kabsch, W. Solution for Best Rotation to Relate 2 Sets of Vectors. Acta Crystallogr. Sect. A 1976, 32, 922–923. [Google Scholar]
- Casasnovas, J.M.; Springer, T.A.; Liu, J.H.; Harrison, S.C.; Wang, J.H. Crystal structure of ICAM-2 reveals a distinctive integrin recognition surface. Nature 1997, 387, 312–315. [Google Scholar]
- Wormald, M.R.; Dwek, R.A. Glycoproteins: Glycan presentation and protein-fold stability. Structure 1999, 7, R155–160. [Google Scholar]
- Joao, H.C.; Scragg, I.G.; Dwek, R.A. Effects of glycosylation on protein conformation and amide proton exchange rates in RNase B. FEBS Lett 1992, 307, 343–346. [Google Scholar]
- Valderrama-Rincon, J.D.; Fisher, A.C.; Merritt, J.H.; Fan, Y.Y.; Reading, C.A.; Chhiba, K.; Heiss, C.; Azadi, P.; Aebi, M.; Delisa, M.P. An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat. Chem. Biol 2012, 8, 434–436. [Google Scholar]
- Mikami, S.; Kobayashi, T.; Yokoyama, S.; Imataka, H. A hybridoma-based in vitro translation system that efficiently synthesizes glycoproteins. J. Biotechnol 2006, 127, 65–78. [Google Scholar]
- Sanchez, E.J.; Lewis, K.M.; Munske, G.R.; Nissen, M.S.; Kang, C. Glycosylation of skeletal calsequestrin: Implications for its function. J. Biol. Chem 2012, 287, 3042–3050. [Google Scholar]
- Zou, G.; Ochiai, H.; Huang, W.; Yang, Q.; Li, C.; Wang, L.X. Chemoenzymatic synthesis and Fcγ receptor binding of homogeneous glycoforms of antibody Fc domain. Presence of a bisecting sugar moiety enhances the affinity of Fc to FcγIIIa receptor. J. Am. Chem. Soc 2011, 133, 18975–18991. [Google Scholar]
- Murakami, M.; Okamoto, R.; Izumi, M.; Kajihara, Y. Chemical synthesis of an erythropoietin glycoform containing a complex-type disialyloligosaccharide. Angew. Chem. Int. Ed. Engl 2012, 51, 3567–3572. [Google Scholar]
- Aussedat, B.; Fasching, B.; Johnston, E.; Sane, N.; Nagorny, P.; Danishefsky, S.J. Total synthesis of the α-subunit of human glycoprotein hormones: Toward fully synthetic homogeneous human follicle-stimulating hormone. J. Am. Chem. Soc 2012, 134, 3532–3541. [Google Scholar]
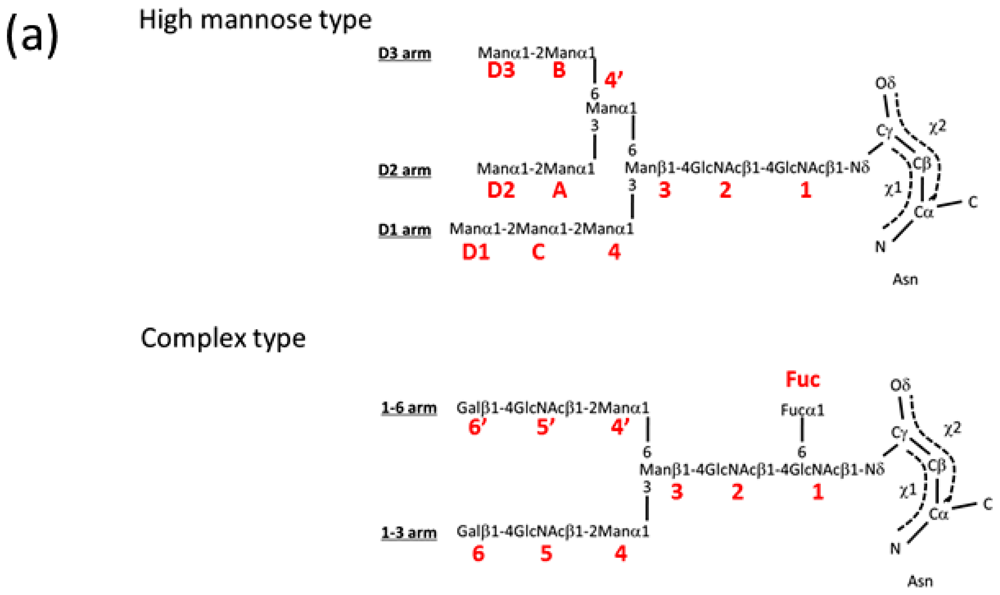
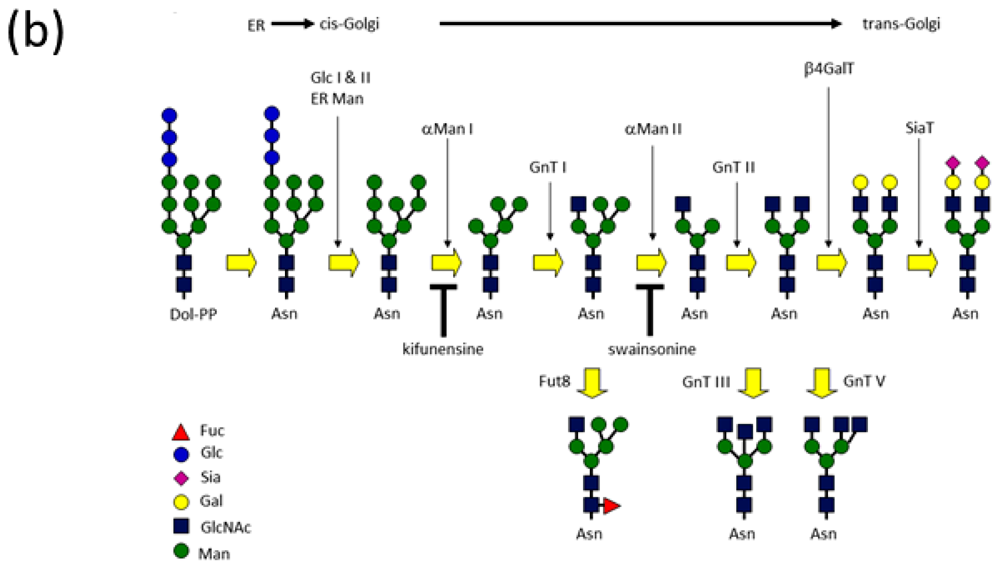

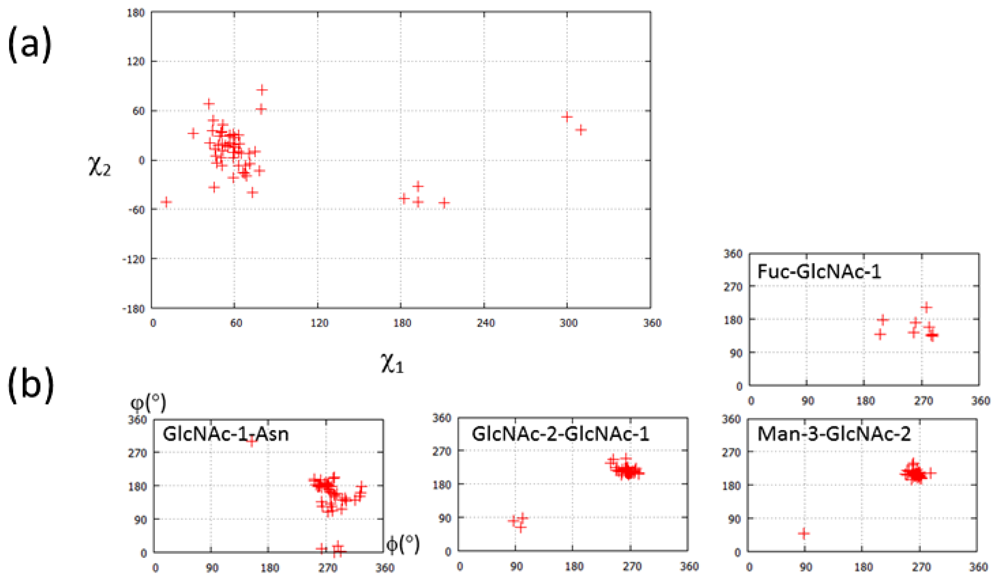
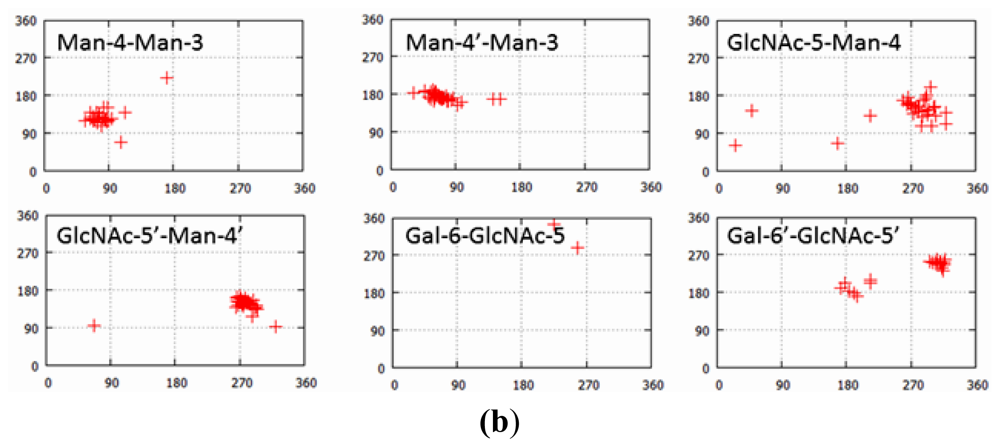

| PDB ID | Glycan structure * | Resolution | Reference |
|---|---|---|---|
| Human IgG1 Fc fragment produced by HEK293T cell + kifunensine | |||
| 2wah | M7GN2 (chain-A) | 2.51 | [30] |
| M1GN2 (chain-B) | - | - | |
| Human IgG1 Fc fragment by enzymatic treatment | |||
| 1h3t | M1GN2F | 2.4 | [31] |
| 1h3u | M3GN2F | 2.4 | enzymatic |
| 1h3x | GN2M3GN2F | 2.44 | treatment |
| 1h3v | G2GN2M3GN2F | 3.1 | [32]) |
| 1h3w | G2GN2M3GN2F | 2.82 | - |
| 1h3y | G2GN2M3GN2F | 4.1 | - |
| Human IgG1 Fc produced by CHO or Fut8−/− CHO cells | |||
| 3ave (2dtq) | GN2M3GN2F | 2.0 | [29] |
| 2dts | GN1M3GN2 (chain-A) | 2.2 | - |
| GN2M3GN2 (chain-B) | - | - | |
| Human IgG1 Fc fragment triple mutant (M252Y/S254T/T256E) | |||
| 3fjt | GN2M3GN2F | 2.5 | [33] |
| Human IgG1 Fc fragment triple mutant (L234F/L235E/P331S) | |||
| 3c2s | G1GN2M3GN2F | 2.3 | [34] |
| Protein is produced by HEK293 cells. | |||
| Human IgG1 Fc fragment triple mutant (S239D/A330L/I332E) | |||
| 2ql1 | G1GN2M3GN2F | 2.5 | [35] |
| Human IgG1 Fc fragment + 13 residues peptide | |||
| 1dn2 | G1GN1M3GN2F (chain-A) | 2.7 | [36] |
| G1GN2M3GN2F (chain-B) | - | - | |
| Human IgG1 (Rituxan) Fc fragment + Staphylococcus aureus Protein A domain B | |||
| 1l6x | G2GN2M3GN2F | 1.65 | [37] |
| Human IgG1 Fc fragment | |||
| 3do3 | G1GN2M3GN2F | 2.5 | [38] |
| Human IgG1 | |||
| 1hzh | G2GN2M3GN2F (chain-H) | 2.7 | [39] |
| G1GN2M3GN2F (chain-K) | - | - | |
| Mouse IgG2a | |||
| 1igt | G1GN2M3GN2F | 2.8 | [19] |
| Human IgG1 Fc fragment + Protein-A mimetic peptide dendrimer. | |||
| 3d6g | GN2M3GN2F | 2.30 | [40] |
| Mouse IgG2b Fc fragment | |||
| 2rgs | GN2M3GN2F | 2.1 | [41] |
| Rabbit IgG Fc fragment | |||
| 2vuo | G1GN2M3GN2 | 1.95 | [42] |
| Human IgG1 Fc fragment + minimized protein A | |||
| 1oqo | GN2M3GN2F | 2.3 | [43] |
| 1oqx | M3GN2F | 2.6 | - |
| Human IgG Fc fragment | |||
| 1fc1 | G1GN2M3GN2F | 2.9 | [17] |
| Rat IgG2a Fc fragment | |||
| 1i1c | GN2M3GN2F | 2.7 | [18] |
| Human IgG1 Fc fragment + human Fc receptor (FcγRIIIb) | |||
| 1t83 (1its) | G1GN1M3GN2F (chain-A) | 3.0 | [44] |
| GN2M3GN2F (chain-B) | - | - | |
| 1t89 (1iix) | G1GN1M3GN2F (chain-A) | 3.5 | - |
| GN2M3GN2F (chain-B) | - | - | |
| Human IgG1 Fc fragment + human Fc receptor (FcγRIIIb) | |||
| 1e4k | G1GN2M3GN2F | 3.2 | [45] |
| Human IgG1 Fc fragment + human Fc receptor (FcγRIIIa) | |||
| 3sgj | GN2M3GN2F | 2.2 | [46] |
| 3sgk | GN3M3GN2 | 2.4 | - |
| Human IgG1 Fc fragment + human Fc receptor (FcγRIIIa) | |||
| 3ay4 | G1GN2M3GN2 | 2.2 | [47] |
| Human heterodimeric Fc + human neonatal FcR (FnRn) | |||
| 1i1a | GN2M3GN2F | 2.8 | [18] |
| PDB ID | Glycan structure * | Resolution | Reference |
|---|---|---|---|
| N2 (A/Tokyo/3/1967) | |||
| 1nn2 | M4GN2 | 2.20 | [74] |
| N2 (A/Tokyo/3/1967) | |||
| 1inw | M3GN2 | 2.40 | [75] |
| 1inx | M3GN2 | 2.40 | - |
| N6 (A/swine/KU/2/2001) | |||
| 1v0z | M3-5GN2 | 1.84 | [76] |
| 1w1x | M1-5GN2 | 2.00 | - |
| 1w20 | M1-6GN2 | 2.08 | - |
| 1w21 | M1-6GN2 | 2.08 | - |
| 2cml | M6GN2 | 2.15 | - |
| N9 (A/Tern/Australia/G70C/75) | |||
| 1iny | M5GN2 | 2.40 | [75] |
| N9 (A/Tern/Australia/G70C/1975 (H11N9)) | |||
| 1f8b | M5GN2 | 1.80 | [77] |
| 1f8c | M5GN2 | 1.70 | - |
| 1f8d | M5GN2 | 1.40 | - |
| 1f8e | M5GN2 | 1.40 | - |
| N9 (A/Tern/Australia/G70C/1975) in complex with single chain Fv fragment | |||
| 1a14 | M5GN2 | 2.50 | [78] |
| N9 (A/NWS/whale/Maine/1/84) | |||
| 2b8h | M7-8GN2 | 2.20 | [79] |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nagae, M.; Yamaguchi, Y. Function and 3D Structure of the N-Glycans on Glycoproteins. Int. J. Mol. Sci. 2012, 13, 8398-8429. https://doi.org/10.3390/ijms13078398
Nagae M, Yamaguchi Y. Function and 3D Structure of the N-Glycans on Glycoproteins. International Journal of Molecular Sciences. 2012; 13(7):8398-8429. https://doi.org/10.3390/ijms13078398
Chicago/Turabian StyleNagae, Masamichi, and Yoshiki Yamaguchi. 2012. "Function and 3D Structure of the N-Glycans on Glycoproteins" International Journal of Molecular Sciences 13, no. 7: 8398-8429. https://doi.org/10.3390/ijms13078398
APA StyleNagae, M., & Yamaguchi, Y. (2012). Function and 3D Structure of the N-Glycans on Glycoproteins. International Journal of Molecular Sciences, 13(7), 8398-8429. https://doi.org/10.3390/ijms13078398






