Clinical Relevance of CDH1 and CDH13 DNA-Methylation in Serum of Cervical Cancer Patients
Abstract
:1. Introduction
2. Results and Discussion
2.1. Specificity and Sensitivity of CDH1 and CDH13 DNA-Methylation in Serum Samples
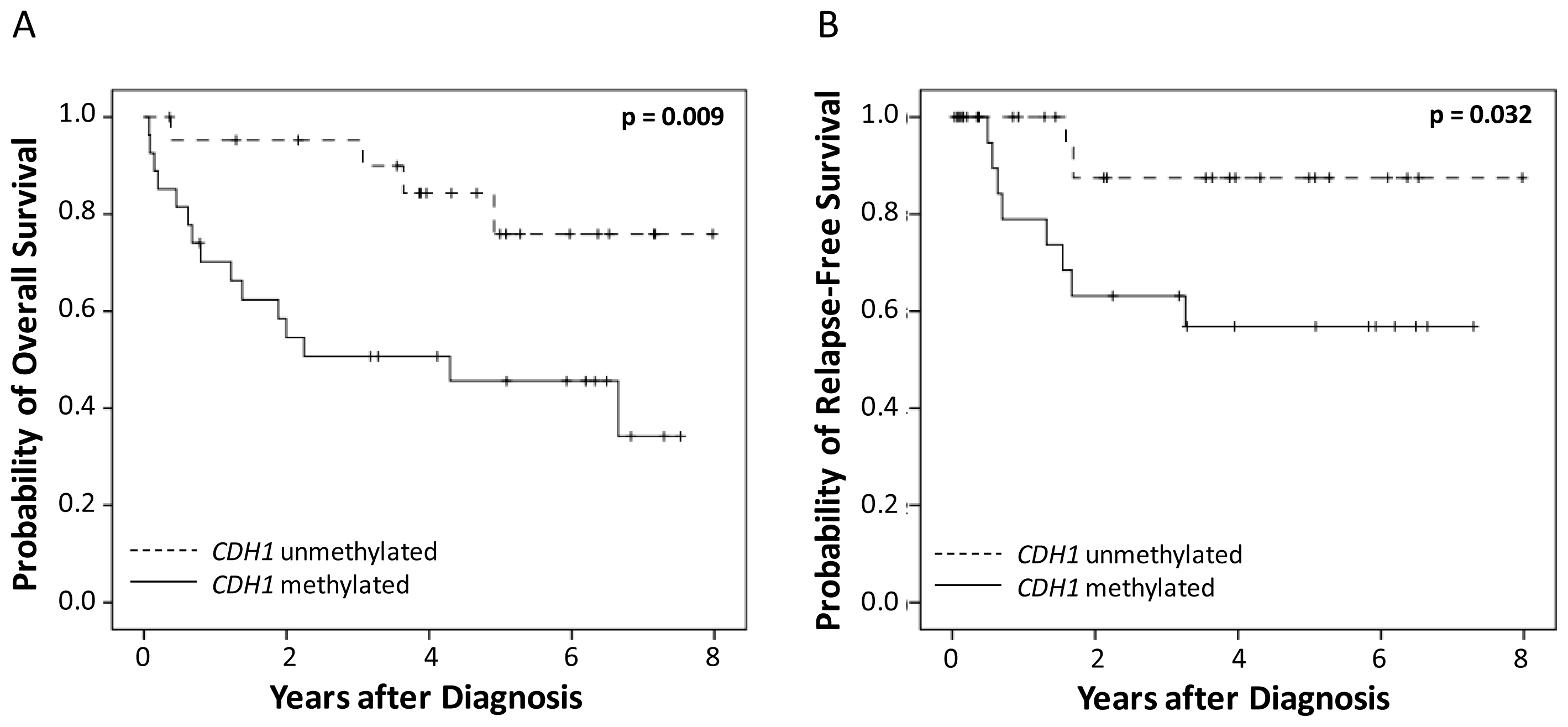
2.2. Correlation of CDH1 and CDH13 DNA-Methylation and Survival
2.2.1. Univariate Survival Analysis
2.2.2. Multivariate Survival Analysis
2.3. Comparison of MethyLight PCR and DHPLC-PCR
2.4. Discussion
3. Experimental Section
3.1. Patients
3.2. Serum Characteristics
3.3. DNA Isolation and MethyLight PCR Analysis
3.4. Denaturing High-Performance Liquid Chromatography Analysis (DHPLC) PCR
3.5. Statistical Analysis
4. Conclusions
Acknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Tost, J. DNA methylation: An introduction to the biology and the disease-associated changes of a promising biomarker. Mol. Biotechnol 2010, 44, 71–81. [Google Scholar]
- Yang, H.J.; Liu, V.W.S.; Wang, Y.; Chan, K.Y.K.; Tsang, P.C.K.; Khoo, U.S.; Cheung, A.N.Y.; Ngan, H.Y.S. Detection of hypermethylated genes in tumor and plasma of cervical cancer patients. Gynecol. Oncol 2004, 93, 435–440. [Google Scholar]
- Widschwendter, A.; Gattringer, C.; Ivarsson, L.; Fiegl, H.; Schneitter, A.; Ramoni, A.; Müller, H.M.; Wiedemair, A.; Jerabek, S.; Müller-Holzner, E.; et al. Analysis of aberrant DNA methylation and human papillomavirus DNA in cervico-vaginal specimens to detect invasive cervical cancer and its precursors. Clin. Cancer Res 2004, 10, 3396–3400. [Google Scholar]
- Widschwendter, A.; Müller, H.M.; Fiegl, H.; Ivarsson, L.; Wiedemair, A.; Müller-Holzner, E.; Goebel, G.; Marth, C.; Widschwendter, M. DNA methylation in serum and tumors of cervical cancer patients. Clin. Cancer Res 2004, 10, 565–571. [Google Scholar]
- Wu, T.; Giovannucci, E.; Welge, J.; Mallick, P.; Tang, W.Y.; Ho, S.M. Measurement of GSTP1 promoter methylation in body fluids may complement PSA screening: A meta-analysis. Br. J. Cancer 2011, 105, 65–73. [Google Scholar]
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med 2011, 9, 133. [Google Scholar]
- Ng, E.K.; Leung, C.P.; Shin, V.Y.; Wong, C.L.; Ma, E.S.; Jin, H.C.; Chu, K.M.; Kwong, A. Quantitative analysis and diagnostic significance of methylated SLC19A3 DNA in the plasma of breast and gastric cancer patients. PLoS One 2011, 6, e22233. [Google Scholar]
- Liggett, T.E.; Melnikov, A.; Yi, Q.; Replogle, C.; Hu, W.; Rotmensch, J.; Kamat, A.; Sood, A.K.; Levenson, V. Distinctive DNA methylation patterns of cell-free plasma DNA in women with malignant ovarian tumors. Gynecol. Oncol 2011, 120, 113–120. [Google Scholar]
- Ostrow, K.L.; Hoque, M.O.; Loyo, M.; Brait, M.; Greenberg, A.; Siegfried, J.M.; Grandis, J.R.; Gaither Davis, A.; Bigbee, W.L.; Rom, W.; et al. Molecular analysis of plasma DNA for the early detection of lung cancer by quantitative methylation-specific PCR. Clin. Cancer Res 2010, 16, 3463–3472. [Google Scholar]
- Oda, H.; Takeichi, M. Evolution: Structural and functional diversity of cadherin at the adherens junction. J. Cell Biol 2011, 193, 1137–1146. [Google Scholar]
- Kuphal, S.; Martyn, A.C.; Pedley, J.; Crowther, L.M.; Bonazzi, V.F.; Parsons, P.G.; Bosserhoff, A.K.; Hayward, N.K.; Boyle, G.M. H-cadherin expression reduces invasion of malignant melanoma. Pigment Cell Melanoma Res 2009, 22, 296–306. [Google Scholar]
- Ling, Z.Q.; Li, P.; Ge, M.H.; Zhao, X.; Hu, F.J.; Fang, X.H.; Dong, Z.M.; Mao, W.M. Hypermethylation-modulated down-regulation of CDH1 expression contributes to the progression of esophageal cancer. Int. J. Mol. Med 2011, 27, 625–635. [Google Scholar]
- Chen, C.L.; Liu, S.S.; Ip, S.-M.; Wong, L.C.; Ngan, H.Y.S. E-cadherin expression is silenced by DNA methylation in cervical cancer cell lines and tumours. Eur. J. Cancer 2003, 39, 517–523. [Google Scholar]
- Sebova, K.; Zmetakova, I.; Bella, V.; Kajo, K.; Stankovicova, I.; Kajabov, V.; Krivulcik, T.; Lasabova, Z.; Tomka, M.; Galbavy, S.; et al. RASSF1A and CDH1 hypermethylation as potential epimarkers in breast cancer. Cancer Biomark 2011–2012, 10, 13–26. [Google Scholar]
- Widschwendter, A.; Ivarsson, L.; Blassnig, A.; Müller, H.M.; Fiegl, H.; Wiedemair, A.; Müller-Holzner, E.; Goebel, G.; Marth, C.; Widschwendter, M. CDH1 and CDH13 methylation in serum is an independent prognostic marker in cervical cancer patients. Int. J. Cancer 2004, 109, 163–166. [Google Scholar]
- Graflund, M.; Sorbe, B.; Hussein, A.; Bryne, M.; Karlsson, M. The prognostic value of histopathologic grading parameters and microvessel density in patients with early squamous cell carcinoma of the uterine cervix. Int. J. Gynecol. Cancer 2002, 12, 32–41. [Google Scholar]
- Metindir, J.; Bilir, G. Prognostic factors affecting disease-free survival in early-stage cervical cancer patients undergoing radical hysterectomy and pelvic-paraaortic lymphadenectomy. Eur. J. Gynaecol. Oncol 2007, 28, 28–32. [Google Scholar]
- Kang, S.; Kim, J.W.; Kang, G.H.; Park, N.H.; Song, Y.S.; Kang, S.B.; Lee, H.P. Polymorphism in folate- and methionine-metabolizing enzyme and aberrant CpG island hypermethylation in uterine cervical cancer. Gynecol. Oncol 2005, 96, 173–180. [Google Scholar]
- Narayan, G.; Arias-Pulido, H.; Koul, S.; Vargas, H.; Zhang, F.F.; Villella, J.; Schneider, A.; Terry, M.B.; Mansukhani, M.; Murty, V.V. Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: Its relationship to clinical outcome. Mol. Cancer 2003, 2, 24. [Google Scholar]
- Wentzensen, N.; Sherman, M.E.; Schiffman, M.; Wang, S.S. Utility of methylation markers in cervical cancer early detection: Appraisal of the state-of-the-science. Gynecol. Oncol 2009, 112, 293–299. [Google Scholar]
- Eads, C.A.; Danenberg, K.D.; Kawakami, K.; Saltz, L.B.; Blake, C.; Shibata, D.; Danenberg, P.V.; Laird, P.W. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000, 28, e32. [Google Scholar]
| MethyLight DHPLC PCR | MethyLight | ||||
|---|---|---|---|---|---|
| Characteristics | No. of Cases (n = 49) | CDH1 (%) * | CDH13 (%) * | No. of Cases (n = 49) | CDH1 (%) * |
| Stage | |||||
| FIGO I | 19 | 10 (53%) | 3 (16%) | 5 (26%) | 3 (16%) |
| FIGO II | 13 | 8 (62%) | 0 (0%) | 3 (23%) | 4 (31%) |
| FIGO III | 12 | 5 (42%) | 1 (8%) | 3 (25%) | 0 (0%) |
| FIGO IV | 5 | 4 (80%) | 1 (20%) | 2 (40%) | 0 (0%) |
| Tumor grade | |||||
| I | 6 | 3 (50%) | 0 (0%) | 1 (17%) | 0 (0%) |
| II | 31 | 18 (58%) | 5 (16%) | 9 (29%) | 5 (16%) |
| III | 12 | 6 (50%) | 0 (0%) | 3 (25%) | 2 (17%) |
| Histology | |||||
| Squamous cell carcinoma | 41 | 21 (51%) | 4 (10%) | 9 (22%) | 4 (10%) |
| Small cell carcinoma | 8 | 6 (75%) | 1 (13%) | 4 (50%) | 3 (38%) |
| Age (years) | |||||
| <50 | 11 | 5 (45%) | 0 (0%) | 4 (36%) | 2 (18%) |
| ≥50 | 38 | 22 (58%) | 5 (13%) | 9 (24%) | 5 (13%) |
| Overall survival | Relapse-free survival | |||
|---|---|---|---|---|
| Variables | No. patients | p value | No. patients | p value |
| (died/total) | (logrank-test) | (relapsed/total) | (logrank-test) | |
| Age | ||||
| <50 | 3/11 | 0.332 | 2/11 | 0.513 |
| ≥50 | 16/38 | 8/38 | ||
| Stage | ||||
| FIGO I | 6/19 | <0.001 | 2/19 | <0.001 |
| FIGO II | 3/13 | 2/13 | ||
| FIGO III | 5/12 | 5/12 | ||
| FIGO IV | 5/5 | 1/5 | ||
| Tumor grade | ||||
| I | 0/6 | 0.127 | 0/6 | 0.058 |
| II | 13/31 | 5/31 | ||
| III | 6/12 | 5/12 | ||
| Surgery | ||||
| no | 16/37 | 0.180 | 8/37 | 0.339 |
| yes | 3/12 | 2/12 | ||
| Chemotherapy | ||||
| no | 13/23 | 0.010 | 4/23 | 0.991 |
| yes | 6/26 | 6/26 | ||
| Radiation therapy | ||||
| no | 4/10 | 0.586 | 1/10 | 0.375 |
| yes | 14/38 | 9/38 | ||
| CDH1 DNA-methylation | ||||
| negative | 4/22 | 0.009 | 2/22 | 0.032 |
| positive | 15/27 | 8/27 | ||
| CDH13 DNA-methylation | ||||
| negative | 17/44 | 0.777 | 9/44 | 0.679 |
| positive | 2/5 | 1/5 | ||
| Overall survival | Relapse-free survival | |||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Age | ||||
| <50 vs. ≥50 | - * | - | - * | - |
| Stage | ||||
| FIGO I/II vs. III/IV | 6.4 (2.1–19.1) | 0.001 | 24.6 (3.5–175.2) | 0.001 |
| Tumor grade | ||||
| I/II vs. III | - * | - | 9.0 (1.5–54.1) | 0.016 |
| Chemotherapy | ||||
| no vs. yes | 0.2 (0.1–0.5) | 0.002 | 0.2 (0.03–1.6) | 0.130 |
| CDH1 DNA-methylation | ||||
| negative vs. positive | 7.8 (2.2–27.7) | 0.001 | 92.8 (3.9–2207.1) | 0.005 |
| CDH13 DNA-methylation | ||||
| negative vs. positive | 2.3 (0.4–12.3) | 0.3 | - * | - |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Abudukadeer, A.; Bakry, R.; Goebel, G.; Mutz-Dehbalaie, I.; Widschwendter, A.; Bonn, G.K.; Fiegl, H. Clinical Relevance of CDH1 and CDH13 DNA-Methylation in Serum of Cervical Cancer Patients. Int. J. Mol. Sci. 2012, 13, 8353-8363. https://doi.org/10.3390/ijms13078353
Abudukadeer A, Bakry R, Goebel G, Mutz-Dehbalaie I, Widschwendter A, Bonn GK, Fiegl H. Clinical Relevance of CDH1 and CDH13 DNA-Methylation in Serum of Cervical Cancer Patients. International Journal of Molecular Sciences. 2012; 13(7):8353-8363. https://doi.org/10.3390/ijms13078353
Chicago/Turabian StyleAbudukadeer, Abida, Rania Bakry, Georg Goebel, Irene Mutz-Dehbalaie, Andreas Widschwendter, Günther K. Bonn, and Heidi Fiegl. 2012. "Clinical Relevance of CDH1 and CDH13 DNA-Methylation in Serum of Cervical Cancer Patients" International Journal of Molecular Sciences 13, no. 7: 8353-8363. https://doi.org/10.3390/ijms13078353




