Risk-Association of DNA Methyltransferases Polymorphisms with Gastric Cancer in the Southern Chinese Population
Abstract
:1. Introduction
2. Results and Discussion
2.1. Subject Characteristics
2.2. Genotype Information
2.3. MAF of the Chosen SNPs with HapMap Data
2.4. Associations between Individual SNPs and GC Risk
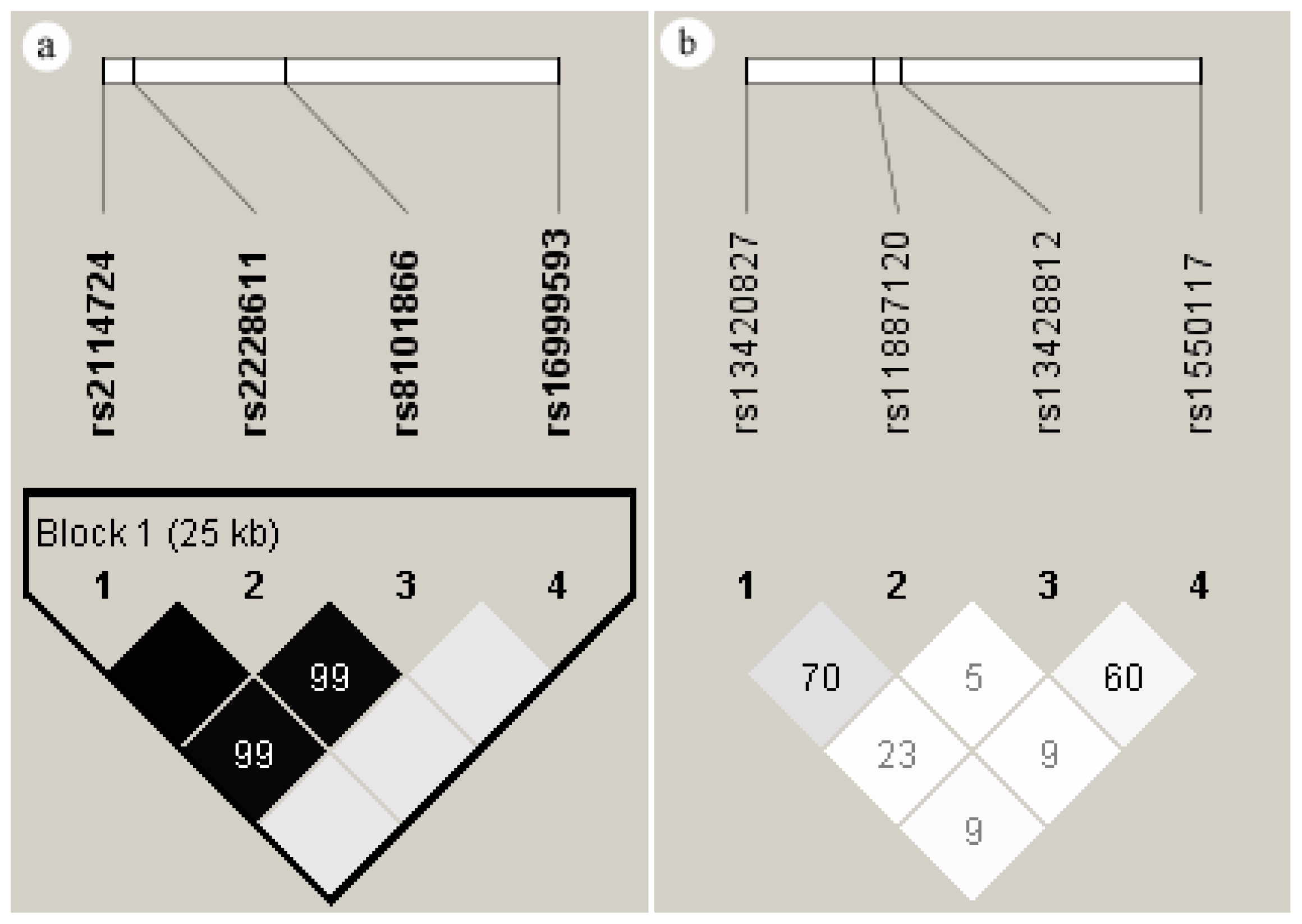
2.5. Linkage Disequilibrium of the SNPs in DNMT1 and DNMT3A
2.6. Haplotypes of the SNPs in DNMT1 and DNMT3A
3. Experimental Section
3.1. Subjects
3.2. SNP Selection and Genotyping
3.3. Data Quality Assessment and Statistical Analysis
4. Conclusions
Supplementary Information
ijms-13-08364-s001.pdfAcknowledgment
- Conflict of InterestThe authors declare no conflict of interest.
References
- Yang, L. Incidence and mortality of gastric cancer in China. World J. Gastroenterol 2006, 12, 17–20. [Google Scholar]
- Leung, W.K.; Wu, M.S.; Kakugawa, Y.; Kim, J.J.; Yeoh, K.G.; Goh, K.L.; Wu, K.C.; Wu, D.C.; Sollano, J.; Kachintorn, U.; et al. Screening for gastric cancer in Asia: Current evidence and practice. Lancet Oncol 2008, 9, 279–287. [Google Scholar]
- Cebrian, A.; Pharoah, P.D.; Ahmed, S.; Ropero, S.; Fraga, M.F.; Smith, P.L.; Conroy, D.; Luben, R.; Perkins, B.; Easton, D.F.; et al. Genetic variants in epigenetic genes and breast cancer risk. Carcinogenesis 2006, 27, 1661–1669. [Google Scholar]
- Bheemanaik, S.; Reddy, Y.V.; Rao, D.N. Structure, function and mechanism of exocyclic DNA methyltransferases. Biochem. J 2006, 399, 177–190. [Google Scholar]
- Goll, M.G.; Bestor, T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem 2005, 74, 481–514. [Google Scholar]
- Liu, Y.; Oakeley, E.J.; Sun, L.; Jost, J.P. Multiple domains are involved in the targeting of the mouse DNA methyltransferase to the DNA replication foci. Nucleic Acids Res 1998, 26, 1038–1045. [Google Scholar]
- Schaefer, M.; Lyko, F. Solving the DNMT2 enigma. Chromosoma 2010, 119, 35–40. [Google Scholar]
- Mizuno, S.; Chijiwa, T.; Okamura, T.; Akashi, K.; Fukumaki, Y.; Niho, Y.; Sasaki, H. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood 2001, 97, 1172–1179. [Google Scholar]
- Li, E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet 2002, 3, 662–673. [Google Scholar]
- Wienholz, B.L.; Kareta, M.S.; Moarefi, A.H.; Gordon, C.A.; Ginno, P.A.; Chedin, F. DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet 2010, 6, e1001106. [Google Scholar]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases DNMT3A and DNMT3B are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar]
- Bourc’his, D.; Bestor, T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking DNMT3L. Nature 2004, 431, 96–99. [Google Scholar]
- Kaneda, M.; Okano, M.; Hata, K.; Sado, T.; Tsujimoto, N.; Li, E.; Sasaki, H. Essential role for de novo DNA methyltransferase DNMT3A in paternal and maternal imprinting. Nature 2004, 429, 900–903. [Google Scholar]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet 2005, 6, 597–610. [Google Scholar]
- Delaval, K.; Wagschal, A.; Feil, R. Epigenetic deregulation of imprinting in congenital diseases of aberrant growth. Bioessays 2006, 28, 453–459. [Google Scholar]
- Feinberg, A.P.; Tycko, B. The history of cancer epigenetics. Nat. Rev. Cancer 2004, 4, 143–153. [Google Scholar]
- Balassiano, K.; Lima, S.; Jenab, M.; Overvad, K.; Tjonneland, A.; Boutron-Ruault, M.C.; Clavel-Chapelon, F.; Canzian, F.; Kaaks, R.; Boeing, H.; et al. Aberrant DNA methylation of cancer-associated genes in gastric cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST). Cancer Lett 2011, 311, 85–95. [Google Scholar]
- El-Maarri, O.; Kareta, M.S.; Mikeska, T.; Becker, T.; Diaz-Lacava, A.; Junen, J.; Nusgen, N.; Behne, F.; Wienker, T.; Waha, A.; et al. A systematic search for DNA methyltransferase polymorphisms reveals a rare DNMT3L variant associated with subtelomeric hypomethylation. Hum. Mol. Genet 2009, 18, 1755–1768. [Google Scholar]
- Liu, K.; Wang, Y.F.; Cantemir, C.; Muller, M.T. Endogenous assays of DNA methyltransferases: Evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol. Cell Biol 2003, 23, 2709–2719. [Google Scholar]
- Kanai, Y.; Ushijima, S.; Ochiai, A.; Eguchi, K.; Hui, A.; Hirohashi, S. DNA hypermethylation at the D17S5 locus is associated with gastric carcinogenesis. Cancer Lett 1998, 122, 135–141. [Google Scholar]
- Khatami, F.; Noorinayer, B.; Ghiasi, S.; Mohebi, R.; Hashemi, M.; Zali, M.R. Lack of effects of single nucleotide polymorphisms of the DNA methyltransferase 1 gene on gastric cancer in Iranian patients: A case control study. Asian Pac. J. Cancer Prev 2009, 10, 1177–1182. [Google Scholar]
- Fan, H.; Liu, D.; Qiu, X.; Qiao, F.; Wu, Q.; Su, X.; Zhang, F.; Song, Y.; Zhao, Z.; Xie, W. A functional polymorphism in the DNA methyltransferase-3A promoter modifies the susceptibility in gastric cancer but not in esophageal carcinoma. BMC Med 2010, 8, 12. [Google Scholar]
- Hu, J.; Fan, H.; Liu, D.; Zhang, S.; Zhang, F.; Xu, H. DNMT3B promoter polymorphism and risk of gastric cancer. Dig. Dis. Sci 2010, 55, 1011–1016. [Google Scholar]
- Aung, P.P.; Matsumura, S.; Kuraoka, K.; Kunimitsu, K.; Yoshida, K.; Matsusaki, K.; Nakayama, H.; Yasui, W. No evidence of correlation between the single nucleotide polymorphism of DNMT3B promoter and gastric cancer risk in a Japanese population. Oncol. Rep 2005, 14, 1151–1154. [Google Scholar]
- NCBI SNP Database. Available online: http://www.ncbi.nlm.nih.gov/projects/SNP/ accessed on 13 October 2011.
- Xiang, G.; Zhenkun, F.; Shuang, C.; Jie, Z.; Hua, Z.; Wei, J.; Da, P.; Dianjun, L. Association of DNMT1 gene polymorphisms in exons with sporadic infiltrating ductal breast carcinoma among Chinese Han women in the Heilongjiang Province. Clin. Breast Cancer 2010, 10, 373–377. [Google Scholar]
- Kelemen, L.E.; Sellers, T.A.; Schildkraut, J.M.; Cunningham, J.M.; Vierkant, R.A.; Pankratz, V.S.; Fredericksen, Z.S.; Gadre, M.K.; Rider, D.N.; Liebow, M.; et al. Genetic variation in the one-carbon transfer pathway and ovarian cancer risk. Cancer Res 2008, 68, 2498–2506. [Google Scholar]
- Kelemen, L.E.; Goodman, M.T.; McGuire, V.; Rossing, M.A.; Webb, P.M.; Kobel, M.; Anton-Culver, H.; Beesley, J.; Berchuck, A.; Brar, S.; et al. Genetic variation in TYMS in the one-carbon transfer pathway is associated with ovarian carcinoma types in the Ovarian Cancer Association Consortium. Cancer Epidemiol. Biomark. Prev 2010, 19, 1822–1830. [Google Scholar]
- Paz, M.F.; Fraga, M.F.; Avila, S.; Guo, M.; Pollan, M.; Herman, J.G.; Esteller, M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res 2003, 63, 1114–1121. [Google Scholar]
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004, 429, 457–463. [Google Scholar]
- Wjst, M. Target SNP selection in complex disease association studies. BMC Bioinform 2004, 5, 92. [Google Scholar]
- Hermann, A.; Goyal, R.; Jeltsch, A. The DNMT1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem 2004, 279, 48350–48359. [Google Scholar]
- Chen, T.; Hevi, S.; Gay, F.; Tsujimoto, N.; He, T.; Zhang, B.; Ueda, Y.; Li, E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet 2007, 39, 391–396. [Google Scholar]
- Mutze, K.; Langer, R.; Schumacher, F.; Becker, K.; Ott, K.; Novotny, A.; Hapfelmeier, A.; Hofler, H.; Keller, G. DNA methyltransferase 1 as a predictive biomarker and potential therapeutic target for chemotherapy in gastric cancer. Eur. J. Cancer 2011, 47, 1817–1825. [Google Scholar]
- Yang, J.; Wei, X.; Wu, Q.; Xu, Z.; Gu, D.; Jin, Y.; Shen, Y.; Huang, H.; Fan, H.; Chen, J. Clinical significance of the expression of DNA methyltransferase proteins in gastric cancer. Mol. Med. Rep 2011, 4, 1139–1143. [Google Scholar]
- Ding, W.J.; Fang, J.Y.; Chen, X.Y.; Peng, Y.S. The expression and clinical significance of DNA methyltransferase proteins in human gastric cancer. Dig. Dis. Sci 2008, 53, 2083–2089. [Google Scholar]
- Etoh, T.; Kanai, Y.; Ushijima, S.; Nakagawa, T.; Nakanishi, Y.; Sasako, M.; Kitano, S.; Hirohashi, S. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am. J. Pathol 2004, 164, 689–699. [Google Scholar]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog DNMT2. Science 2006, 311, 395–398. [Google Scholar]
- Hermann, A.; Schmitt, S.; Jeltsch, A. The human DNMT2 has residual DNA-(cytosine-C5) methyltransferase activity. J. Biol. Chem 2003, 278, 31717–31721. [Google Scholar]
- Ushijima, T.; Sasako, M. Focus on gastric cancer. Cancer Cell 2004, 5, 121–125. [Google Scholar]
- Samuel, M.S.; Suzuki, H.; Buchert, M.; Putoczki, T.L.; Tebbutt, N.C.; Lundgren-May, T.; Christou, A.; Inglese, M.; Toyota, M.; Heath, J.K.; et al. Elevated DNMT3A activity promotes polyposis in Apc(Min) mice by relaxing extracellular restraints on Wnt signaling. Gastroenterology 2009, 137. [Google Scholar]
- Watanabe, Y.; Kim, H.S.; Castoro, R.J.; Chung, W.; Estecio, M.R.; Kondo, K.; Guo, Y.; Ahmed, S.S.; Toyota, M.; Itoh, F.; et al. Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes. Gastroenterology 2009, 136, 2149–2158. [Google Scholar]
- Wang, Y.M.; Wang, R.; Wen, D.G.; Li, Y.; Guo, W.; Wang, N.; Wei, L.Z.; He, Y.T.; Chen, Z.F.; Zhang, X.F.; et al. Single nucleotide polymorphism in DNA methyltransferase 3B promoter and its association with gastric cardiac adenocarcinoma in North China. World J. Gastroenterol 2005, 11, 3623–3627. [Google Scholar]
- Fan, H.; Liu, D.S.; Zhang, S.H.; Hu, J.B.; Zhang, F.; Zhao, Z.J. DNMT3B 579 G>T promoter polymorphism and risk of esophagus carcinoma in Chinese. World J. Gastroenterol 2008, 14, 2230–2234. [Google Scholar]
- Montgomery, K.G.; Liu, M.C.; Eccles, D.M.; Campbell, I.G. The DNMT3B C→T promoter polymorphism and risk of breast cancer in a British population: A case-control study. Breast Cancer Res 2004, 6, R390–R394. [Google Scholar]
- Jones, J.S.; Amos, C.I.; Pande, M.; Gu, X.; Chen, J.; Campos, I.M.; Wei, Q.; Rodriguez-Bigas, M.; Lynch, P.M.; Frazier, M.L. DNMT3B polymorphism and hereditary nonpolyposis colorectal cancer age of onset. Cancer Epidemiol. Biomark. Prev 2006, 15, 886–891. [Google Scholar]
- Chedin, F.; Lieber, M.R.; Hsieh, C.L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by DNMT3A. Proc. Natl. Acad. Sci. USA 2002, 99, 16916–16921. [Google Scholar]
- Kareta, M.S.; Botello, Z.M.; Ennis, J.J.; Chou, C.; Chedin, F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem 2006, 281, 25893–25902. [Google Scholar]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar]
- Gokul, G.; Ramakrishna, G.; Khosla, S. Reprogramming of HeLa cells upon DNMT3L overexpression mimics carcinogenesis. Epigenetics 2009, 4, 322–329. [Google Scholar]
- Haggarty, P.; Hoad, G.; Harris, S.E.; Starr, J.M.; Fox, H.C.; Deary, I.J.; Whalley, L.J. Human intelligence and polymorphisms in the DNA methyltransferase genes involved in epigenetic marking. PLoS One 2010, 5, e11329. [Google Scholar]
- International HapMap Project Database. Available online: http://hapmap.ncbi.nlm.nih.gov/ accessed on 13 June 2011.
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar]
| Variable | Cases (n = 242) N (%) | Controls (n = 294) N (%) | p | |
|---|---|---|---|---|
| Sex | Male | 174 (71.9) | 175 (59.5) | 0.03 a |
| Female | 68 (28.1) | 119 (40.5) | - | |
| Mean (SD) | 54.9 (12.5) | 58.4 (16.4) | 0.005 b | |
| Age (years) | ≤60 | 155 (64.0) | 146 (49.7) | 0.179 |
| >60 | 87 (36.0) | 148 (50.3) | - | |
| Tumor sites | Non-cardiac | 24 (9.9) | - | - |
| Cardiac | 218 (90.1) | - | - | |
| Histological types | Well | 21 (8.7) | - | - |
| Moderate | 42 (17.4) | - | - | |
| Poor | 132 (54.5) | - | - | |
| Signet ring cell | 19 (7.8) | - | - | |
| Unclassified | 28 (11.6) | - | - | |
| Clinical stage (TNM) | I | 19 (7.9) | - | - |
| II | 49 (20.2) | - | - | |
| III | 119 (49.2) | - | - | |
| IV | 55 (22.7) | - | - | |
| Depth of invasion c | T1 | 26 (10.7) | - | - |
| T2 | 64 (26.4) | - | - | |
| T3 | 104 (43.0) | - | - | |
| T4 | 48 (19.8) | - | - | |
| No. | SNP | Gene | Location | From | To | Change | Codon |
|---|---|---|---|---|---|---|---|
| 1 | rs2114724 | DNMT1 | Intron | G | A | 10265248A>G | — |
| 2 | rs2228611 | DNMT1 | Coding region | G | A | Pro463Pro | 463 |
| 3 | rs2228612 | DNMT1 | Coding region | A | C/G/T | Ile327Leu,Phe,Val | 327 |
| 4 | rs8101866 | DNMT1 | Intron | A | G | 10275660G>A | — |
| 5 | rs16999593 | DNMT1 | Coding region | T | C | His97Arg | 97 |
| 6 | rs11254413 | DNMT2 | Coding region | A | G | His101Tyr | 101 |
| 7 | rs6733301 | DNMT3A | Intron | G | A | 25276284G>A | — |
| 8 | rs13420827 | DNMT3A | 3′ untranslated region | G | C | 25453968C>G | — |
| 9 | rs11695471 | DNMT3A | Intron | T | A | 25457708T>A | — |
| 10 | rs11887120 | DNMT3A | Intron | T | A/C/G | 25485735G>T,C,A | — |
| 11 | rs13428812 | DNMT3A | Intron | A | G | 25492467A>G | — |
| 12 | rs1550117 | DNMT3A | 5′ near gene | G | A | 25565907A>G | — |
| 13 | rs6087990 | DNMT3B | 5′ near gene | T | C | 31349908T>C | — |
| 14 | rs2424908 | DNMT3B | Intron | C | T | 31360383C>T | — |
| 15 | rs2424913 | DNMT3B | Intron | C | T | 31374259C>T | — |
| 16 | rs113593938 | DNMT3L | Coding region | C | T | Arg271Gln | 271 |
| Genes/SNP | Minor Allele | Present Study | HapMap Data | ||||
|---|---|---|---|---|---|---|---|
| Control | GC | Beijing | Japan | European | African | ||
| DNMT1 | |||||||
| rs2114724 | T | 0.26 | 0.25 | 0.27 | 0.38 | 0.51 | 0.53 |
| rs2228611 | A | 0.26 | 0.25 | 0.27 | 0.39 | 0.51 | 0.53 |
| rs8101866 | C | 0.19 | 0.23 | 0.27 | 0.44 | 0.53 | 0.52 |
| rs16999593 | C | 0.26 | 0.26 | 0.17 | 0.2 | 0.00 | 0.00 |
| DNMT2 | |||||||
| rs11254413 | A | 0.21 | 0.13 | 0.28 | 0.23 | 0.48 | 0.42 |
| DNMT3A | |||||||
| rs1550117 | A | 0.19 | 0.2 | 0.16 | 0.13 | 0.17 | 0.18 |
| rs11887120 | C | 0.48 | 0.51 | 0.52 | 0.51 | 0.61 | 0.54 |
| rs13420827 | G | 0.2 | 0.18 | 0.24 | 0.3 | 0.3 | 0.33 |
| rs13428812 | G | 0.26 | 0.26 | 0.15 | 0.22 | 0.08 | 0.05 |
| DNMT3B | |||||||
| rs2424908 | C | 0.43 | 0.44 | 0.5 | 0.57 | 0.81 | 0.64 |
| Genes/SNP | Genotype | Cases N (%) | Controls N (%) | Co-Dominant Model | Overdominant Model | ||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p a | OR (95% CI) | p b | ||||
| DNMT1 | |||||||
| rs2114724 | CC | 132 (54.5) | 162 (56.2) | 1.00 | 0.22 | 1.00 | 0.27 |
| CT | 97 (40.1) | 101 (35.1) | 1.16 (0.81–1.68) | - | 1.23 (0.86–1.76) | - | |
| TT | 13 (5.4) | 25 (8.7) | 0.62 (0.30–1.27) | - | - | - | |
| rs2228611 | GG | 132 (54.5) | 160 (56.1) | 1.00 | 0.14 | 1.00 | 0.22 |
| AG | 97 (40.1) | 99 (34.7) | 1.18 (0.81–1.71) | - | 1.26 (0.87–1.80) | - | |
| AA | 13 (5.4) | 26 (9.1) | 0.58 (0.28–1.18) | - | - | - | |
| rs8101866 | TT | 130 (53.9) | 166 (56.5) | 1.00 | 0.17 | 1.00 | 0.19 |
| CT | 98 (40.7) | 102 (34.7) | 1.20 (0.83–1.74) | - | 1.27 (0.89–1.82) | - | |
| CC | 13 (5.4) | 26 (8.8) | 0.61 (0.30–1.26) | - | - | - | |
| rs16999593 | TT | 141 (58.3) | 196 (66.7) | 1.00 | 0.13 | 1.00 | 0.05 |
| CT | 89 (36.8) | 83 (28.2) | 1.47 (1.01–2.14) | - | 1.45 (1.00–2.11) | - | |
| CC | 12 (5.0) | 15 (5.1) | 1.14 (0.51–2.54) | - | - | - | |
| DNMT2 | |||||||
| rs11254413 | GG | 204 (84.3) | 187 (63.8) | 1.00 | <0.0001 | 1.00 | <0.0001 |
| AG | 15 (6.2) | 91 (31.1) | 0.16 (0.09–0.28) | - | 0.15 (0.08–0.27) | - | |
| AA | 23 (9.5) | 15 (5.1) | 1.45 (0.73–2.90) | - | - | - | |
| DNMT3A | |||||||
| rs1550117 | GG | 157 (64.9) | 191 (65.0) | 1.00 | 0.74 | 1.00 | 0.68 |
| AG | 74 (30.6) | 93 (31.6) | 0.94 (0.64–1.37) | - | 0.92 (0.63–1.34) | - | |
| AA | 11 (4.5) | 10 (3.4) | 1.34 (0.55–3.29) | - | - | - | |
| rs11887120 | TT | 57 (23.6) | 74 (25.3) | 1.00 | 0.46 | 1.00 | 0.39 |
| CT | 121 (50.0) | 155 (53.1) | 0.96 (0.63–1.47) | - | 0.86 (0.61–1.22) | - | |
| CC | 64 (26.4) | 63 (21.6) | 1.26 (0.76–2.07) | - | - | - | |
| rs13420827 | CC | 167 (69.0) | 183 (62.7) | 1.00 | 0.046 | 1.00 | 0.034 |
| CG | 61 (25.2) | 99 (33.9) | 0.68 (0.46–1.01) | - | 0.66 (0.45–0.97) | - | |
| GG | 14 (5.8) | 10 (3.4) | 1.75 (0.74–4.14) | - | - | - | |
| rs13428812 | AA | 137 (56.6) | 160 (55.4) | 1.00 | 0.84 | 1.00 | 0.63 |
| AG | 84 (34.7) | 106 (36.7) | 0.93 (0.64–1.35) | - | 0.91 (0.64–1.31) | - | |
| GG | 21 (8.7) | 23 (8.0) | 1.11 (0.58–2.12) | - | - | - | |
| DNMT3B | |||||||
| rs2424908 | TT | 78 (32.2) | 99 (33.7) | 1.00 | 0.96 | 1.00 | 0.82 |
| CT | 114 (47.1) | 139 (47.3) | 0.98 (0.66–1.45) | - | 0.96 (0.68–1.36) | - | |
| CC | 50 (20.7) | 56 (19.1) | 1.05 (0.64–1.71) | - | - | - | |
| Genes | Haplotype | Frequencies | OR * (95% CI) | p Value | ||
|---|---|---|---|---|---|---|
| Total | Control | Case | ||||
| DNMT1 | CGTT | 0.526 | 0.539 | 0.510 | 1.00 | - |
| TACT | 0.257 | 0.258 | 0.254 | 1.01 (0.76–1.36) | 0.93 | |
| CGTC | 0.211 | 0.192 | 0.234 | 1.24 (0.91–1.70) | 0.17 | |
| DNMT3A | CCAG | 0.268 | 0.276 | 0.255 | 1.00 | - |
| CTAG | 0.194 | 0.181 | 0.208 | 1.27 (0.80–2.01) | 0.31 | |
| CCGG | 0.105 | 0.086 | 0.130 | 1.47 (0.82–2.63) | 0.19 | |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yang, X.-X.; He, X.-Q.; Li, F.-X.; Wu, Y.-S.; Gao, Y.; Li, M. Risk-Association of DNA Methyltransferases Polymorphisms with Gastric Cancer in the Southern Chinese Population. Int. J. Mol. Sci. 2012, 13, 8364-8378. https://doi.org/10.3390/ijms13078364
Yang X-X, He X-Q, Li F-X, Wu Y-S, Gao Y, Li M. Risk-Association of DNA Methyltransferases Polymorphisms with Gastric Cancer in the Southern Chinese Population. International Journal of Molecular Sciences. 2012; 13(7):8364-8378. https://doi.org/10.3390/ijms13078364
Chicago/Turabian StyleYang, Xue-Xi, Xuan-Qiu He, Fen-Xia Li, Ying-Song Wu, Yang Gao, and Ming Li. 2012. "Risk-Association of DNA Methyltransferases Polymorphisms with Gastric Cancer in the Southern Chinese Population" International Journal of Molecular Sciences 13, no. 7: 8364-8378. https://doi.org/10.3390/ijms13078364
APA StyleYang, X.-X., He, X.-Q., Li, F.-X., Wu, Y.-S., Gao, Y., & Li, M. (2012). Risk-Association of DNA Methyltransferases Polymorphisms with Gastric Cancer in the Southern Chinese Population. International Journal of Molecular Sciences, 13(7), 8364-8378. https://doi.org/10.3390/ijms13078364




