Oxidative Stress Mediated-Alterations of the MicroRNA Expression Profile in Mouse Hippocampal Neurons
Abstract
:1. Introduction
2. Results
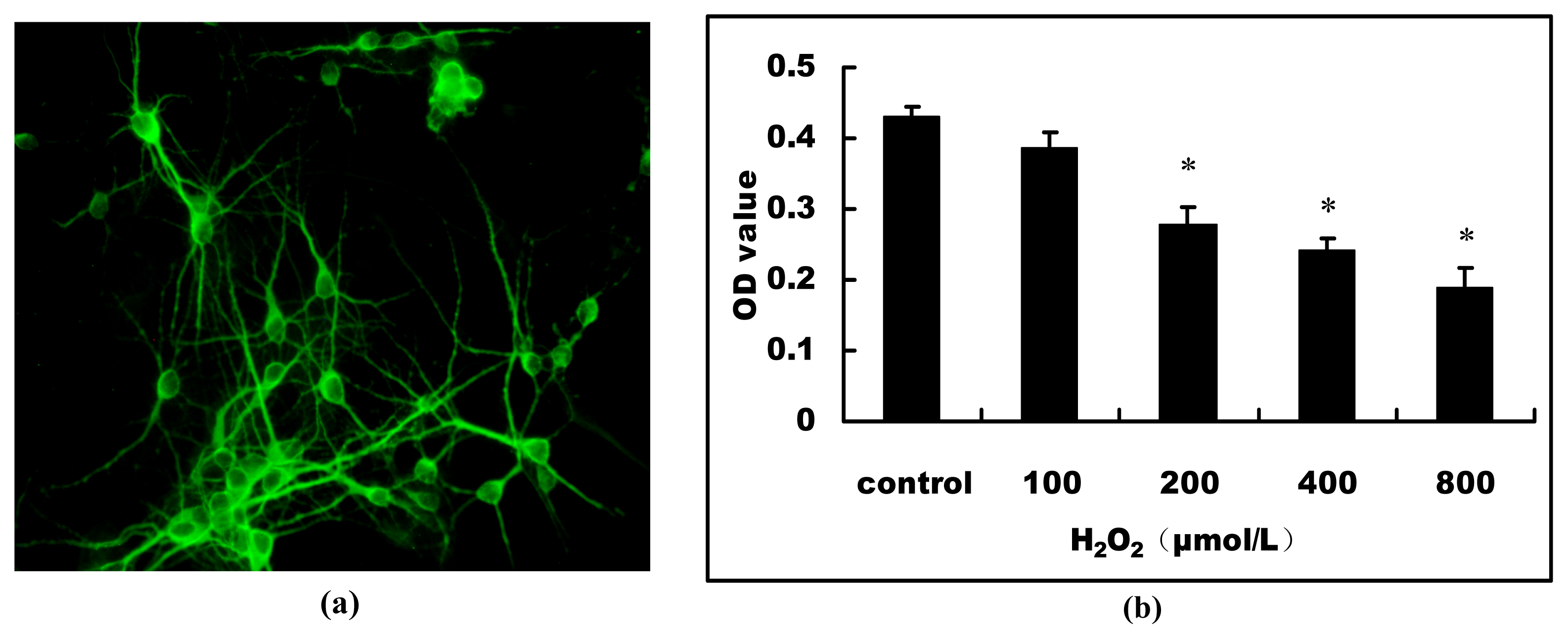
2.1. H2O2-Induced Cell Viability Loss in Primary Hippocampal Neurons
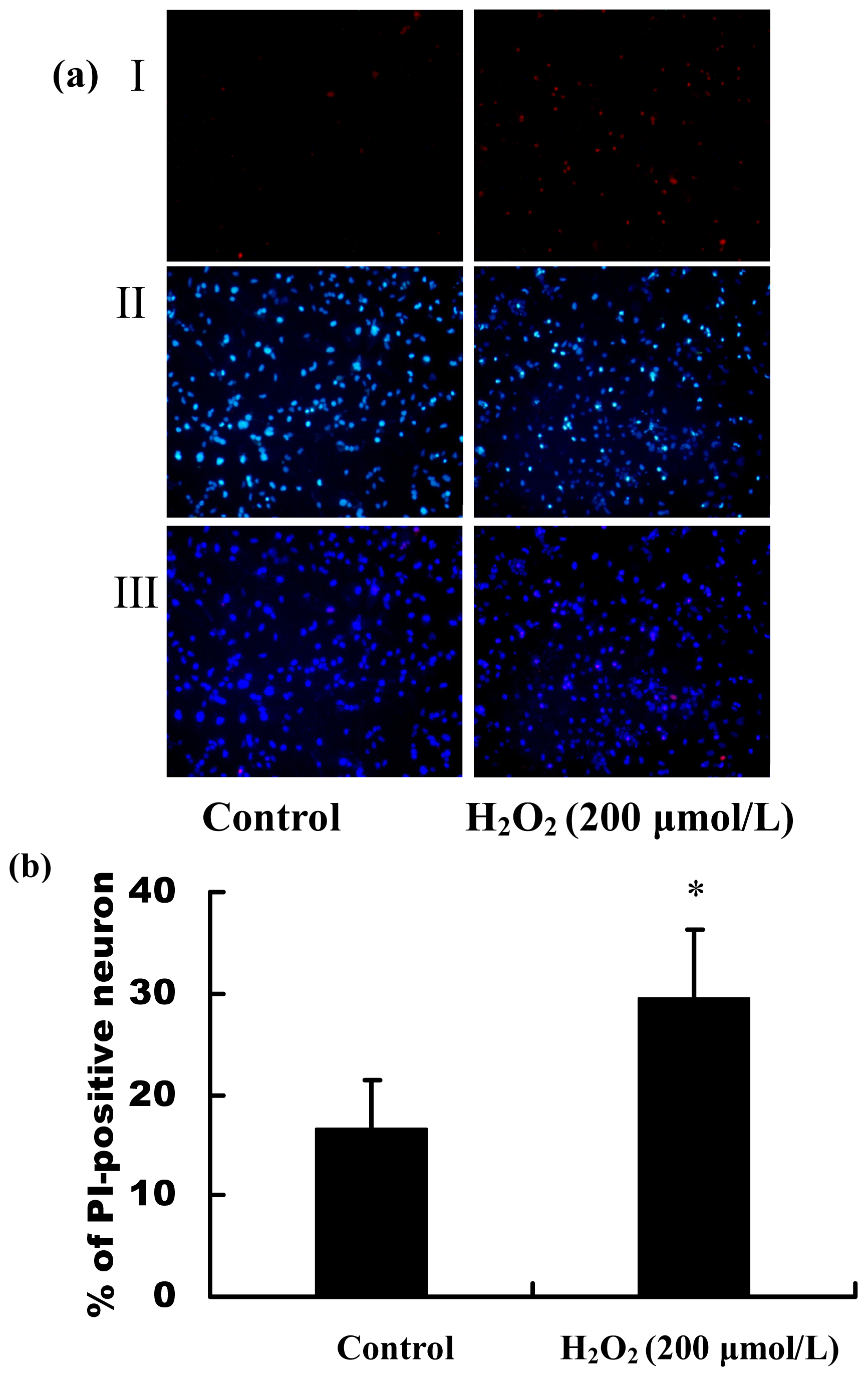
2.2. H2O2 Induced Apoptosis and Death of Primary Cultured Hippocampal Neurons
2.3. Identification of Neuronal miRNAs Modulated by Oxidative Stress
2.4. Deregulated miRNAs May Take Part in Regulating the Key Pathways Altered in AD
2.5. Validation of Deregulated miRNAs
2.6. Functional Analysis of Targets of miR-135b and miR-708
3. Discussion
4. Experimental Section
4.1. Materials
4.2. Cell Culture
4.3. Immunocytochemistry
4.4. Cell Viability
4.5. Measurements of Neuron Apoptosis and Cell Death Induced by H2O2
4.6. Microarray Analysis of miRNA Expression
4.7. Bioinformatic Analysis of Deregulated miRNAs
4.8. Quantitative Real-Time PCR
4.9. Statistical Analysis
5. Conclusions
Acknowledgments
References
- Ramalingam, M.; Kim, S.J. Reactive oxygen/nitrogen species and their functional correlations in neurodegenerative diseases. J. Neural. Transm 2012, 119, 891–910. [Google Scholar]
- Shulman, R.G.; Rothman, D.L.; Behar, K.L.; Hyder, F. Energetic basis of brain activity: Implications for neuroimaging. Trends Neurosci 2004, 27, 489–495. [Google Scholar]
- Su, B.; Wang, X.; Nunomura, A.; Moreira, P.I.; Lee, H.G.; Perry, G.; Smith, M.A.; Zhu, X. Oxidative stress signaling in Alzheimer’s disease. Curr. Alzheimer Res 2008, 5, 525–532. [Google Scholar]
- Braak, H.; Braak, E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol. Aging 1995, 16, 271–284. [Google Scholar]
- Giannakopoulos, P.; Hof, P.R.; Bouras, C. Selective vulnerability of neocortical association areas in Alzheimer’s disease. Microsc. Res. Tech 1998, 43, 16–23. [Google Scholar]
- Colangelo, V.; Schurr, J.; Ball, M.J.; Pelaez, R.P.; Bazan, N.G.; Lukiw, W.J. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: Transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J. Neurosci. Res 2002, 70, 462–473. [Google Scholar]
- Praticò, D. Oxidative stress hypothesis in Alzheimer’s disease: A reappraisal. Trends Pharmacol. Sci 2008, 29, 609–615. [Google Scholar]
- Bishop, N.A.; Lu, T.; Yankner, B.A. Neural mechanisms of ageing and cognitive decline. Nature 2010, 464, 529–535. [Google Scholar]
- Smith, M.A.; Richey, P.L.; Taneda, S.; Kutty, R.K.; Sayre, L.M.; Monnier, V.M.; Perry, G. Advanced Maillard reaction end products, free radicals, and protein oxidation in Alzheimer’s disease. Ann. NY Acad. Sci 1994, 738, 447–454. [Google Scholar]
- Smith, M.A.; Perry, G.; Richey, P.L.; Sayre, L.M.; Anderson, V.E.; Beal, M.F.; Kowall, N. Oxidative damage in Alzheimer’s. Nature 1996, 382, 120–121. [Google Scholar]
- Gibson, G.; Martins, R.; Blass, J.; Gandy, S. Altered oxidation and signal transduction systems in fibroblasts from Alzheimer patients. Life Sci 1996, 59, 477–489. [Google Scholar]
- Curti, D.; Rognoni, F.; Gasparini, L.; Cattaneo, A.; Paolillo, M.; Racchi, M.; Zani, L.; Bianchetti, A.; Trabucchi, M.; Bergamaschi, S. Oxidative metabolism in cultured fibroblasts derived from sporadic Alzheimer’s disease (AD) patients. Neurosci. Lett 1997, 236, 13–16. [Google Scholar]
- Butterfield, D.A.; Lauderback, C.M. Lipid peroxidation and protein oxidation in Alzheimer’s disease brain: Potential causes and consequences involving amyloid β-peptide-associated free radical oxidative stress. Free Radic. Biol. Med 2002, 32, 1050–1060. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar]
- Nunez-Iglesias, J.; Liu, C.C.; Morgan, T.E.; Finch, C.E.; Zhou, X.J. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS One 2010, 5, e8898. [Google Scholar]
- Milton, N.G. Role of hydrogen peroxide in the aetiology of Alzheimer’s disease: Implications for treatment. Drugs Aging 2004, 21, 81–100. [Google Scholar]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar]
- Ramamoorthy, M.; Sykora, P.; Scheibye-Knudsen, M.; Dunn, C.; Kasmer, C.; Zhang, Y.; Becker, K.G.; Croteau, D.L.; Bohr, V.A. Sporadic Alzheimer disease fibroblasts display an oxidative stress phenotype. Free Radic. Biol. Med 2012, 53, 1371–1380. [Google Scholar]
- Chang, T.C.; Mendell, J.T. microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet 2007, 8, 215–239. [Google Scholar]
- Coolen, M.; Bally-Cuif, L. MicroRNAs in brain development and physiology. Curr. Opin. Neurobiol 2009, 19, 461–470. [Google Scholar]
- Yokota, T. MicroRNA and central nervous system. Brain Nerve 2009, 61, 167–176. [Google Scholar]
- Cogswell, J.P.; Ward, J.; Taylor, I.A.; Waters, M.; Shi, Y.; Cannon, B.; Kelnar, K.; Kemppainen, J.; Brown, D.; Chen, C. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis 2008, 14, 27–41. [Google Scholar]
- Shioya, M.; Obayashi, S.; Tabunoki, H.; Arima, K.; Saito, Y.; Ishida, T.; Satoh, J. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol. Appl. Neurobiol 2010, 36, 320–330. [Google Scholar]
- Zovoilis, A.; Agbemenyah, H.Y.; Agis-Balboa, R.C.; Stilling, R.M.; Edbauer, D.; Rao, P.; Farinelli, L.; Delalle, I.; Schmitt, A.; Falkai, P. microRNA-34c is a novel target to treat dementias. EMBO J 2011, 30, 4299–4308. [Google Scholar]
- Leung, A.K.; Sharp, P.A. MicroRNA functions in stress responses. Mol. Cell 2010, 40, 205–215. [Google Scholar]
- Van Dam, D.; Vloeberghs, E.; Abramowski, D.; Staufenbiel, M.; de Deyn, P.P. APP23 mice as a model of Alzheimer’s disease: An example of a transgenic approach to modeling a CNS disorder. CNS Spectr 2005, 10, 207–222. [Google Scholar]
- Patnaik, S.K.; Kannisto, E.; Knudsen, S.; Yendamuri, S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res 2010, 70, 36–45. [Google Scholar]
- Schotte, D.; Chau, J.C.; Sylvester, G.; Liu, G.; Chen, C.; van der Velden, V.H.; Broekhuis, M.J.; Peters, T.C.; Pieters, R.; den Boer, M.L. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia 2009, 23, 313–322. [Google Scholar]
- Saini, S.; Yamamura, S.; Majid, S.; Shahryari, V.; Hirata, H.; Tanaka, Y.; Dahiya, R. MicroRNA-708 induces apoptosis and suppresses tumorigenicity in renal cancer cells. Cancer Res 2011, 71, 6208–6219. [Google Scholar]
- Matsuyama, H.; Suzuki, H.I.; Nishimori, H.; Noguchi, M.; Yao, T.; Komatsu, N.; Mano, H.; Sugimoto, K.; Miyazono, K. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood 2011, 118, 6881–6892. [Google Scholar]
- Lulla, R.R.; Costa, F.F.; Bischof, J.M.; Chou, P.M.; de Fatima Bonaldo, M.; Vanin, E.F.; Soares, M.B. Identification of Differentially Expressed MicroRNAs in Osteosarcoma. Sarcoma 2011, 2011, 732690. [Google Scholar]
- Oberg, A.L.; French, A.J.; Sarver, A.L.; Subramanian, S.; Morlan, B.W.; Riska, S.M.; Borralho, P.M.; Cunningham, J.M.; Boardman, L.A.; Wang, L. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS One 2011, 6, e20465. [Google Scholar]
- Famulski, J.K.; Trivedi, N.; Howell, D.; Yang, Y.; Tong, Y.; Gilbertson, R.; Solecki, D.J. Siah regulation of Pard3A controls neuronal cell adhesion during germinal zone exit. Science 2010, 330, 1834–1838. [Google Scholar]
- Urano, Y.; Iiduka, M.; Sugiyama, A.; Akiyama, H.; Uzawa, K.; Matsumoto, G.; Kawasaki, Y.; Tashiro, F. Involvement of the mouse Prp19 gene in neuronal/astroglial cell fate decisions. J. Biol. Chem 2006, 281, 7498–7514. [Google Scholar]
- Cheng, Y.; Liu, X.; Zhang, S.; Lin, Y.; Yang, J.; Zhang, C. MicroRNA-21 protects against the H2O2-induced injury on cardiac myocytes via its target gene PDCD4. J. Mol. Cell Cardiol 2009, 47, 5–14. [Google Scholar]
- Targetscan. Available online: http://www.targetscan.org accessed on 4 June 2012.
- DAIAN. Available online: http://www.diana.pcbi.upenn.edu/miRGen/v3/miRGen.html accessed on 4 June 2012.
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar]
- SPSS version 16.0; IBM Corporation: Armonk, NY, USA, 2008.


| miRNA | Total targets in mouse | KEGG pathway | Targets in pathway | p-Value | Total counts in hippo-campus [18] |
|---|---|---|---|---|---|
| miR-135b | 979 | Steroid biosynthesis | 4 | 3.05 × 10−2 | 4299 |
| miR-708 | 109 | Neurotrophin signaling pathway | 6 | 2.48 × 10−4 | 9638 |
| MAPK signaling pathway | 7 | 9.66 × 10−4 | |||
| Axon guidance | 5 | 2.63 × 10−3 | |||
| ErbB signaling pathway | 4 | 6.80 × 10−3 | |||
| Chemokine signaling pathway | 5 | 8.47 × 10−3 | |||
| T cell receptor signaling pathway | 4 | 1.56 × 10−2 | |||
| Insulin signaling pathway | 4 | 2.36 × 10−2 | |||
| miR-296 | 777 | MAPK signaling pathway | 23 | 1.57 × 10−4 | 163 |
| Hedgehog signaling pathway | 6 | 4.18 × 10−2 | |||
| Complement and coagulation cascades | 7 | 4.96 × 10−2 | |||
| mir-26b | 948 | Regulation of autophagy | 5 | 4.86 × 10−2 | 227704 |
| miR-201 | 830 | Calcium signaling pathway | 13 | 4.79 × 10−2 | 4 |
| Steroid biosynthesis | 4 | 2.28 × 10−2 | |||
| miR-9 | 888 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 6 | 2.67 × 10−3 | 2298920 |
| ECM-receptor interaction | 8 | 4.69 × 10−2 | |||
| miR-200c | 770 | Neurotrophin signaling pathway | 24 | 9.97 × 10−11 | 178 |
| Insulin signaling pathway | 19 | 1.46 × 10−6 | |||
| Axon guidance | 17 | 1.35 × 10−5 | |||
| MAPK signaling pathway | 25 | 1.88 × 10−5 | |||
| ErbB signaling pathway | 13 | 5.00 × 10−5 | |||
| mTOR signaling pathway | 10 | 1.01 × 10−4 | |||
| Apoptosis | 10 | 3.53 × 10−3 | |||
| Wnt signaling pathway | 11 | 3.88 × 10−2 | |||
| miR-377 | 327 | MAPK signaling pathway | 10 | 1.59 × 10−2 | 1775 |
| Ubiquitin mediated proteolysis | 7 | 1.50 × 10−2 | |||
| TGF-beta signaling pathway | 5 | 3.95 × 10−2 | |||
| mTOR signaling pathway | 4 | 4.53 × 10−2 | |||
| miR-713 | 810 | Spliceosome | 10 | 3.82 × 10−2 | ~ |
| miR-470 | 739 | ECM-receptor interaction | 7 | 4.94 × 10−2 | 4 |
| miR-190b | 805 | Valine, leucine and isoleucine degradation | 6 | 2.68 × 10−2 | 4958 |
| miR-1190 | 215 | MAPK signaling pathway | 9 | 2.10 × 10−3 | ~ |
| VEGF signaling pathway | 4 | 2.95 × 10−2 | |||
| Regulation of actin cytoskeleton | 6 | 4.24 × 10−2 | |||
| miR-32 | 949 | p-value cutoff not met | ~ | ~ | 5042 |
| miR-297 | 895 | No Pathways predicted | ~ | ~ | 13 |
| miR-1956 | 3 | No Pathways predicted | ~ | ~ | 4 |
| miR-369 | ~ | ~ | ~ | ~ | 3556 |
| miR-291a | ~ | ~ | ~ | ~ | ~ |
| miR-135 targets GO-term Analysis | p-Value | |
|---|---|---|
| Biological process | ||
| 1 | GO:0008544~epidermis development | 1.96 × 10−3 |
| 2 | GO:0006310~DNA recombination | 3.62 × 10−3 |
| 3 | GO:0016567~protein ubiquitination | 1.87 × 10−2 |
| 4 | GO:0046777~protein amino acid autophosphorylation | 1.87×10−2 |
| 5 | GO:0002520~immune system development | 2.09×10−2 |
| Cellular component | ||
| 1 | GO:0005829~cytosol | 3.48 × 10−3 |
| 2 | GO:0045177~apical part of cell | 5.01 × 10−3 |
| 3 | GO:0016324~apical plasma membrane | 9.00 × 10−3 |
| 4 | GO:0005681~spliceosome | 3.74 × 10−2 |
| 5 | GO:0044429~mitochondrial part | 3.77 × 10−2 |
| Molecular function | ||
| 1 | GO:0008134~transcription factor binding | 8.07 × 10−3 |
| 2 | GO:0004713~protein tyrosine kinase activity | 1.28 × 10−2 |
| 3 | GO:0043169~cation binding | 1.42 × 10−2 |
| 4 | GO:0043167~ion binding | 1.52 × 10−2 |
| 5 | GO:0046872~metal ion binding | 1.60 × 10−2 |
| miR-708 targets GO-term Analysis | p-Value | |
|---|---|---|
| Biological process | ||
| 1 | GO:0051057~positive regulation of small GTPase mediated signal transduction | 3.42 × 10−3 |
| 2 | GO:0006468~protein amino acid phosphorylation | 4.21 × 10−3 |
| 3 | GO:0045444~fat cell differentiation | 5.53 × 10−3 |
| 4 | GO:0008284~positive regulation of cell proliferation | 6.52 × 10−3 |
| 5 | GO:0048666~neuron development | 7.44 × 10−3 |
| Cellular component | ||
| 1 | GO:0042995~cell projection | 6.54 × 10−3 |
| 2 | GO:0030425~dendrite | 1.65 × 10−2 |
| 3 | GO:0043005~neuron projection | 3.14 × 10−2 |
| 4 | GO:0009898~internal side of plasma membrane | 5.12 × 10−2 |
| 5 | GO:0043228~non-membrane-bounded organelle | 7.54 × 10−2 |
| Molecular function | ||
| 1 | GO:0004674~protein serine/threonine kinase activity | 1.93 × 10−3 |
| 2 | GO:0017076~purine nucleotide binding | 4.22 × 10−3 |
| 3 | GO:0032555~purine ribonucleotide binding | 6.28 × 10−3 |
| 4 | GO:0032553~ribonucleotide binding | 6.28 × 10−3 |
| 5 | GO:0000166~nucleotide binding | 1.04 × 10−2 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Xu, S.; Zhang, R.; Niu, J.; Cui, D.; Xie, B.; Zhang, B.; Lu, K.; Yu, W.; Wang, X.; Zhang, Q. Oxidative Stress Mediated-Alterations of the MicroRNA Expression Profile in Mouse Hippocampal Neurons. Int. J. Mol. Sci. 2012, 13, 16945-16960. https://doi.org/10.3390/ijms131216945
Xu S, Zhang R, Niu J, Cui D, Xie B, Zhang B, Lu K, Yu W, Wang X, Zhang Q. Oxidative Stress Mediated-Alterations of the MicroRNA Expression Profile in Mouse Hippocampal Neurons. International Journal of Molecular Sciences. 2012; 13(12):16945-16960. https://doi.org/10.3390/ijms131216945
Chicago/Turabian StyleXu, Shunjiang, Rui Zhang, Jingya Niu, Dongsheng Cui, Bing Xie, Binggui Zhang, Kang Lu, Wenjun Yu, Xueyi Wang, and Qingfu Zhang. 2012. "Oxidative Stress Mediated-Alterations of the MicroRNA Expression Profile in Mouse Hippocampal Neurons" International Journal of Molecular Sciences 13, no. 12: 16945-16960. https://doi.org/10.3390/ijms131216945




