Rapid Development of Microsatellite Markers for the Endangered Fish Schizothorax biddulphi (Günther) Using Next Generation Sequencing and Cross-Species Amplification
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sequencing by Ion Torrent PGM™
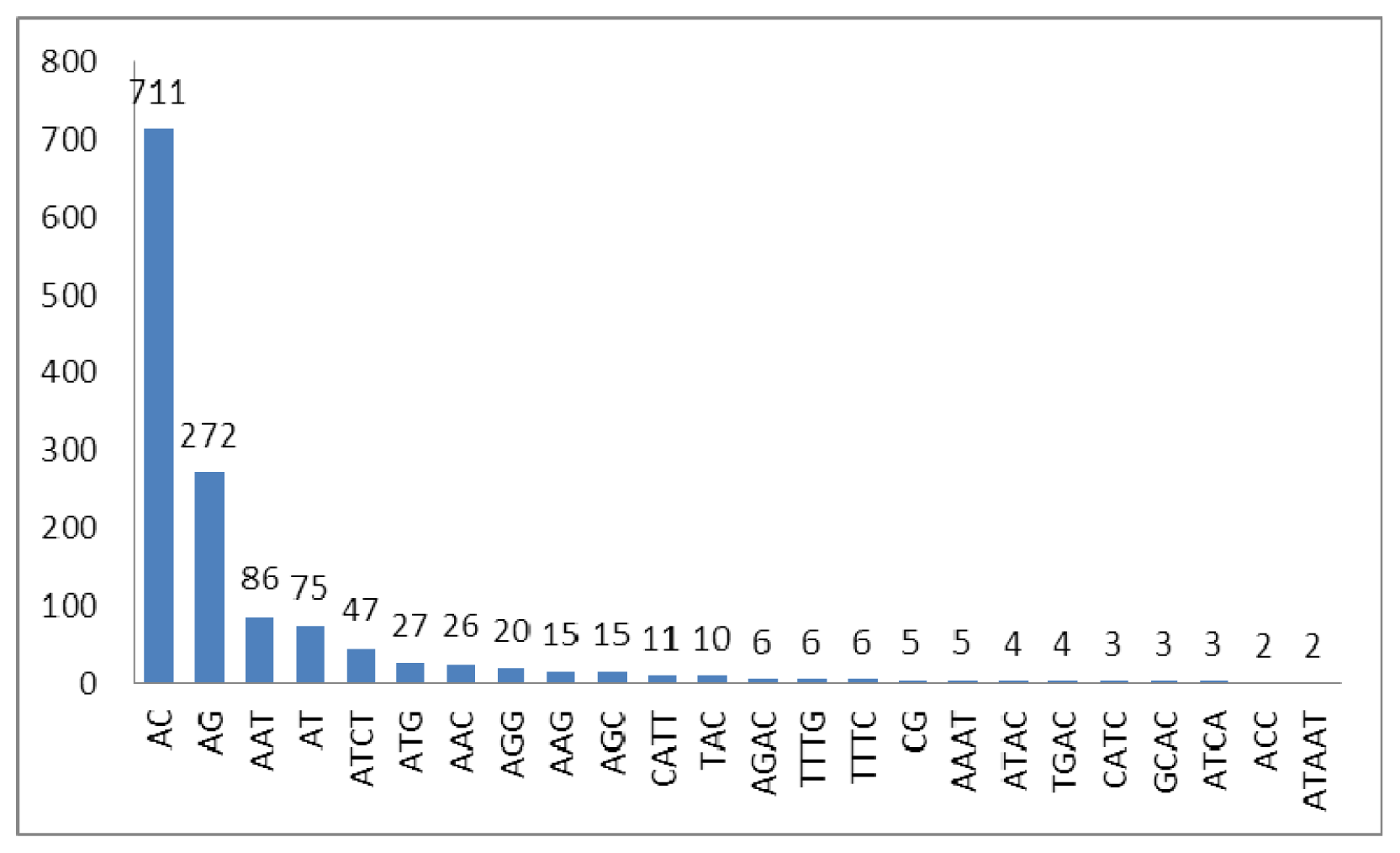
2.2. Characteristics of Microsatellites
2.3. SSR Polymorphism
2.4. Cross-Amplification in S. irregularis
3. Experimental Section
3.1. Sample and Genetic DNA Extraction
3.2. Ion Torrent PGM™ Library Preparation and Sequencing
3.3. Mining SSR Loci and Primer Design
3.4. PCR Amplification and Genotyping
3.5. Data Analysis
3.6. Microsatellite Markers Cross-Amplification in S. irregularis
4. Conclusions
Acknowledgments
References
- Zhang, R.M.; Guo, Y.; Ma, Y.W. Turxun (2007) A survey on the resource and distribution of Schizothorax biddulphi Günther. Freshw. Fish 2007, 37, 76–78. [Google Scholar]
- Liu, X.J.; Hu, G.F. Threatened fishes of the world: Schizothorax (Schizopyge) biddulphi Günther, 1876 (Cyprinidae). Environ. Biol. Fish 2009, 85, 97–98. [Google Scholar]
- Yue, P.Q.; Chen, Y.Y. China Red Databook of Endangered Animals (Pisce.); Science Press: Beijing, China, 1998; pp. 153–155. [Google Scholar]
- Mariette, S.; Le Corre, V.; Austerlitz, F.; Kremer, A. Sampling within the genome for measuring within-population diversity: Tradeoffs between markers. Mol. Ecol 2002, 11, 1145–1156. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol 2005, 14, 2611–2620. [Google Scholar]
- Castoe, T.A.; Poole, A.W.; Gu, W.; Koning, A.P.J.D.; Daza, J.M.; Smith, E.N.; Pollock, D.D. Rapid identification of thousands of copperhead snake (Agkistrodon contortrix) microsatellite loci from modest amounts of 454 shotgun genome sequence. Mol. Ecol. Resour 2010, 10, 341–347. [Google Scholar]
- Abdelkrim, J.; Robertson, B.C.; Stanton, J.-A.L.; Gemmell, N.J. Fast, cost-effective development of species-specific microsatellite markers by genomic sequencing. Biotechniques 2009, 46, 185–192. [Google Scholar]
- Gardner, M.G.; Fitch, A.J.; Bertozzi, T.; Lowe, A.J. Rise of the machines—Recommendations for ecologists when using next generation sequencing for microsatellite development. Mol. Ecol. Resour 2011, 11, 1093–1101. [Google Scholar]
- Loman, N.J.; Misra, R.V.; Dallman, T.J.; Constantinidou, C.; Gharbia, S.E.; Wain, J.; Pallen, M.J. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol 2012, 30, 434–439. [Google Scholar] [Green Version]
- Whiteley, A.S.; Jenkins, S.; Waite, I.; Kresoje, N.; Payne, H.; Mullan, B.; Allcock, R.; O’Donnell, A. Microbial 16S rRNA Ion Tag and community metagenome sequencing using the Ion Torrent (PGM) Platform. J. Microbiol. Meth 2012, 91, 81–88. [Google Scholar]
- Somridhivej, B.; Wang, S.; Sha, Z.; Liu, H.; Quilang, J.; Xu, P.; Li, P.; Hu, Z.; Liu, Z. Characterization, polymorphism assessment, and database construction for microsatellites from BAC end sequences of channelcatfish (Ictalurus punctatus): A resource for integration of linkage and physical maps. Aquaculture 2008, 275, 76–80. [Google Scholar]
- Meglécz, E.; Nève, G.; Biffin, E.; Gardner, M.G. Breakdown of phylogenetic signal: A survey of microsatellite densities in 454 shotgun sequences from 154 non-model eukaryote species. PLoS One 2012, 7, e40861. [Google Scholar]
- Edwardsa, Y.J.K.; Elgara, G.; Clarka, M.S.; Bishop, M.J. The identification and characterization of microsatellites in the compact genome of the Japanese pufferfish, Fugu rubripes: Perspectives in functional and comparative genomic analyses. J. Mol. Biol 1998, 278, 843–854. [Google Scholar]
- Saarinen, E.V.; Austin, J.D. When technology meets conservation: Increased microsatellite marker production using 454 genome sequencing on the endangered okaloosa darter (Etheostoma okaloosae). J. Hered 2010, 101, 784–788. [Google Scholar]
- Wang, Y.; Guo, X. Development and Characterization of EST-SSR Markers in the Eastern Oyster Crassostrea virginica. Mar. Biotechnol 2007, 9, 500–511. [Google Scholar]
- Zhan, A.B.; Bao, Z.M.; Wang, X.L.; Hu, J.J. Microsatellite markers derived from bay scallop Argopecten irradians expressed sequence tags. Fish. Sci 2005, 71, 1341–1346. [Google Scholar]
- Katti, M.V.; Ranjekar, P.K.; Gupta, V.S. Differential distribution of simple sequence repeats in eukaryotic fenome sequences. Mol. Biol. Evol 2001, 18, 1161–1167. [Google Scholar]
- Tóth, G.; Gáspári, Z.; Jurka, J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res 2000, 10, 967–981. [Google Scholar]
- Schorderet, D.F.; Gartler, S.M. Analysis of CpG suppression in methylated and nonmethylated species. Proc. Natl. Acad. Sci. USA 1992, 89, 957–961. [Google Scholar]
- Perry, J.C.; Rowe, L. Rapid Microsatellite Development for Water Striders by Next-Generation Sequencing. J. Hered 2011, 102, 125–129. [Google Scholar]
- Csencsics, D.; Brodbeck, S.; Holderegger, R. Cost-effective, species-specific microsatellite development for the endangered Dwarf Bulrush (Typha minima) using next-generation sequencing technology. J. Hered 2010, 101, 789–793. [Google Scholar]
- Chabot, C.L.; Nigenda, S. Characterization of 13 microsatellite loci for the tope shark, Galeorhinus galeus, discovered with next-generation sequencing and their utility for eastern Pacific smooth-hound sharks (Mustelus). Conservation Genet. Resour 2011, 3, 553–555. [Google Scholar]
- Gong, X.; Cui, Z.; Wang, C. Isolation and characterization of polymorphic microsatellite loci from the endangered Tarim schizothoracin (Schizothorax biddulphi Günther). Conserv. Genet. Resour 2012, 4, 795–797. [Google Scholar]
- Liu, X.; Luo, W.; Zeng, C.; Wang, W.; Gao, Z. Isolation of new 40 microsatellite markers in mandarin fish (Siniperca chuatsi). Int. J. Mol. Sci 2011, 12, 4180–4189. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2002; pp. 479–483. [Google Scholar]
- You, F.M.; Huo, N.; Gu, Y.Q.; Luo, M.; Ma, Y.; Hane, D.; Lazo, G.R.; Dvorak, J.; Anderson, O.D. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinformatics 2008, 9, 253. [Google Scholar]
- Jurka, J.; Pethiyagoda, C. Simple repetitive DNA sequences from primates: Compilation and Analysis. J. Mol. Evol 1995, 40, 120–126. [Google Scholar]
- Yeh, F.C.; Yang, R.C.; Boyle, T. POPGENE, version 1.32; Molecular Biology and Biotechnology Centre, University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetic software for exact tests and ecumenicism. J. Hered 1995, 86, 248–249. [Google Scholar]
- Van oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotype errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar]
| Sequencing | Total number of bases (Mbp) | 892.72 |
| Total number of reads | 3,476,226 | |
| Mean length of all reads (bp) | 257 | |
| Longest read (bp) | 399 | |
| Assembly | Total number of contigs after assembly | 40,577 |
| Mean length of contigs (bp) | 395 | |
| Repeat | Number of loci identified | Percentage (%) | Frequency (%) | Mean distance (kb) |
|---|---|---|---|---|
| Dinucleotide | 1063 | 77.08 | 2.62 | 15.11 |
| Trinucleotide | 201 | 14.58 | 0.50 | 79.89 |
| Tetranucleotide | 103 | 7.47 | 0.25 | 155.90 |
| Pentanucleotide | 9 | 0.65 | 0.02 | 1784.16 |
| Hexanucleotide | 3 | 0.22 | 0.01 | 5352.48 |
| Total SSRs | 1379 | 100 | 3.40 | 11.64 |
| Step | Run time (h) | Cost (dollar) | Instructures or softwares |
|---|---|---|---|
| Ion Torrent adapter-ligated library preparation | 7–11 | $100 | Common molecular biology equipment |
| Sample emulsion PCR and enrichment | 4–6.5 | $150 | One Touch V26 |
| Sequencing with a 318 chip | 4.5–5.5 | $600 | Ion Torrent V2.0 |
| Sequence assembly | 5–8 | $100 | CLC Genomics Workbench 5 |
| SSR Mining and primer design | 5–8 | free | BatchPrimer 3 software |
| Total | 25–39 | $950 |
| Locus/GenBank Accession No. | Primer sequence(5′→3′) | Ta (°C) | Repeat motif | Size range (bp) | Na | Ho | He | PHWE | Na in S. irregularis |
|---|---|---|---|---|---|---|---|---|---|
| SCH1/JX473024 | F: GCCATCCTTCAGTTGTGTCT | 62 | (TATC)7 | 240–288 | 6 | 0.70 | 0.83 | 0.00 * | 4 |
| R: AACCGAGTTTCATCCTCCTT | |||||||||
| SCH2/JX473025 | F: CTATGCTCGGTTTCTTTTCA | 57 | (CA)13 | 130–144 | 3 | 0.40 | 0.59 | 0.04 * | 2 |
| R: ACTGATGTGTGTGTGCGTGT | |||||||||
| SCH3/JX473026 | F: ATCCACGCTCTCACACTCTT | 59 | (GT)26 | 192–214 | 3 | 0.75 | 0.54 | 0.15 | 1 |
| R: CCAGCTCCTCAACACAGATG | |||||||||
| SCH4/JX473027 | F: GTGTGTGTGTGCGAGAGTGT | 54 | (TG)10 | 211–231 | 3 | 0.60 | 0.59 | 0.11 | 2 |
| R: TTCAGATGTAACCCCCTTTG | |||||||||
| SCH5/JX473028 | F: TGAAAGTTCCTTTGCTCCTG | 52 | (TG)10 | 188–234 | 3 | 0.70 | 0.62 | 0.00 * | 2 |
| R: GTGACACACTGTGCAAAAGC | |||||||||
| SCH6/JX473029 | F: GTGTGTGTGTGTGTGTGTGTG | 59 | (TC)12 | 130–156 | 4 | 0.75 | 0.66 | 0.17 | - |
| R: CCATTACGCCTATGGAATGT | |||||||||
| SCH7/JX473030 | F: GTGGGGTGATGGAAAATACA | 59 | (GT)11 | 190–222 | 3 | 0.75 | 0.62 | 0.56 | 1 |
| R: GGCTGACCATTGTGCTAAAC | |||||||||
| SCH8/JX473031 | F: AAGGTTGAACAGTTGTTTGC | 54 | (TTA)7 | 105–125 | 3 | 0.75 | 0.61 | 0.04 * | - |
| R: ATGTCCAGTGTAGCGACTGA | |||||||||
| SCH9/JX473032 | F: GTGCAGCTCTGTCTCGATCT | 57 | (TG)13 | 218–240 | 3 | 0.35 | 0.52 | 0.17 | - |
| R: TGTGGATTGTTGCAGTGTTT | |||||||||
| SCH10/JX473033 | F: TTCATTGTTGCATTCCTTCC | 51 | (TTCC)5 | 192–234 | 3 | 0.75 | 0.61 | 0.07 | 3 |
| R: GTTGGTGATGGTGTTCTGCT | |||||||||
| SCH11/JX473034 | F: CGGCAACCAGACCGTGTA | 50 | (GT)15 | 190–214 | 4 | 0.50 | 0.52 | 0.51 | 2 |
| R: CTCCCATACCGCTCCTCC | |||||||||
| SCH12/JX473035 | F: TAAAATCGAAGGGGAACA | 50 | (TG)15 | 151–183 | 3 | 0.70 | 0.66 | 0.12 | 2 |
| R:GACAGTGAGAAGAGGAAACA | |||||||||
| SCH13/JX473036 | F: TTTCCCCTTAGTCATTTC | 50 | (AG)10 | 100–124 | 3 | 0.55 | 0.62 | 0.03 * | - |
| R: GGTGTTTGTCAGGAGTTG | |||||||||
| SCH14/JX473037 | F: TTATCTGGACGGAGTGAA | 50 | (AAC)8 | 189–215 | 4 | 0.70 | 0.70 | 0.88 | 2 |
| R: CATTTTGGGGTGAACTAT | |||||||||
| SCH15/JX473038 | F: TCGGTCAATGATGGTGTT | 54 | (ATA)8 | 238–268 | 4 | 0.80 | 0.67 | 0.58 | - |
| R: TTTGGCAGGTCCTTCTTA | |||||||||
| SCH16/JX473039 | F: CACAGATAAGAACACGAAT | 50 | (CA)23 | 242–288 | 3 | 0.15 | 0.27 | 0.02 * | 1 |
| R: AGGGTTTGGAAGAGGTA | |||||||||
| SCH17/JX473040 | F: ACTATTTGTGAGCAGCCC | 59 | (GA)15 | 343–369 | 3 | 0.58 | 0.57 | 0.42 | - |
| R: TATGCGGAAAACCGTGAC | |||||||||
| SCH18/JX473041 | F: TCAATGAGCAACGAAAGAGC | 52 | (AGGCAG)5 | 136–176 | 4 | 0.75 | 0.63 | 0.00 * | 1 |
| R: ATGGTGGCGAAGGGAGAA | |||||||||
| SCH19/JX473042 | F: ACACTCCTGCTACGGTCA | 57 | (TGA)5 | 446–482 | 3 | 0.42 | 0.68 | 0.10 | 1 |
| R: TACATCGCCTCTGCTCCT | |||||||||
| SCH20/JX473043 | F: CGCCAGCGTCTGCCACAA | 58 | (AGC)5 | 220–316 | 6 | 0.64 | 0.75 | 0.11 | 4 |
| R: GCCGCCATCTTCACCCAC | |||||||||
| SCH21/JX473044 | F: TGCCTCAAGGAACTGGTG | 50 | (CGACG)5 | 154–174 | 2 | 0.39 | 0.39 | 0.97 | 2 |
| R: GAGCATTAGAGTATCGTGGT | |||||||||
| SCH22/JX473045 | F: CCGTGGTAAGCACAAGAG | 54 | (CAAA)5 | 340–362 | 3 | 0.63 | 0.48 | 0.30 | 2 |
| R: GACAGCAGGAGGAGAAGG | |||||||||
| SCH23/JX473046 | F: TGACGGTAGAGTCCAGTG | 50 | (CAATTC)5 | 162–184 | 2 | 0.74 | 0.50 | 0.03 * | - |
| R: TGTAATGACGAACAAGCA | |||||||||
| SCH24/JX473047 | F: GACACTGCGTTTTGAAGG | 50 | (AG)11 | 146–168 | 3 | 0.72 | 0.60 | 0.33 | - |
| R: GTCTAACCAGTCGCTCCA | |||||||||
| SCH25/JX473048 | F: CCCAGTTACAGCCTTCTC | 46 | (AC)12 | 195–217 | 2 | 0.30 | 0.51 | 0.06 | 1 |
| R: CAGTTAGTTAGTAGGATGCG | |||||||||
| SCH26/JX473049 | F: ACACTAATAAGCATCAGCAG | 56 | (AC)10 | 147–175 | 5 | 0.50 | 0.64 | 0.26 | 1 |
| R: CACAGTTCACAAGAGCAAG | |||||||||
| SCH28/JX473051 | F: ATGAGAGCAGAAGAGTGGG | 50 | (TG)10 | 262–334 | 4 | 0.85 | 0.71 | 0.58 | 2 |
| R: GAGGAGGCTGTGAGGAAC | |||||||||
| SCH29/JX473052 | F: TTAGAAGTGGAGACAGTT | 56 | (TG)11 | 260–334 | 6 | 0.85 | 0.78 | 0.05 | 3 |
| R: TGAGAGTAAAGAGAGAGC | |||||||||
| SCH30/JX473053 | F: TACTTTCTATCGTGTTTTTG | 48 | (TA)8 | 148–164 | 4 | 0.45 | 0.46 | 0.83 | - |
| R: GTAACCTGCTGAACTTTG | |||||||||
| Mean value | 3.5 | 0.611 | 0.598 | 2.4 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Luo, W.; Nie, Z.; Zhan, F.; Wei, J.; Wang, W.; Gao, Z. Rapid Development of Microsatellite Markers for the Endangered Fish Schizothorax biddulphi (Günther) Using Next Generation Sequencing and Cross-Species Amplification. Int. J. Mol. Sci. 2012, 13, 14946-14955. https://doi.org/10.3390/ijms131114946
Luo W, Nie Z, Zhan F, Wei J, Wang W, Gao Z. Rapid Development of Microsatellite Markers for the Endangered Fish Schizothorax biddulphi (Günther) Using Next Generation Sequencing and Cross-Species Amplification. International Journal of Molecular Sciences. 2012; 13(11):14946-14955. https://doi.org/10.3390/ijms131114946
Chicago/Turabian StyleLuo, Wei, Zhulan Nie, Fanbin Zhan, Jie Wei, Weimin Wang, and Zexia Gao. 2012. "Rapid Development of Microsatellite Markers for the Endangered Fish Schizothorax biddulphi (Günther) Using Next Generation Sequencing and Cross-Species Amplification" International Journal of Molecular Sciences 13, no. 11: 14946-14955. https://doi.org/10.3390/ijms131114946




