A New Strategy for Identification of Highly Conserved microRNAs in Non-Model Insect, Spodoptera litura
Abstract
:1. Introduction
2. Results and Discussion
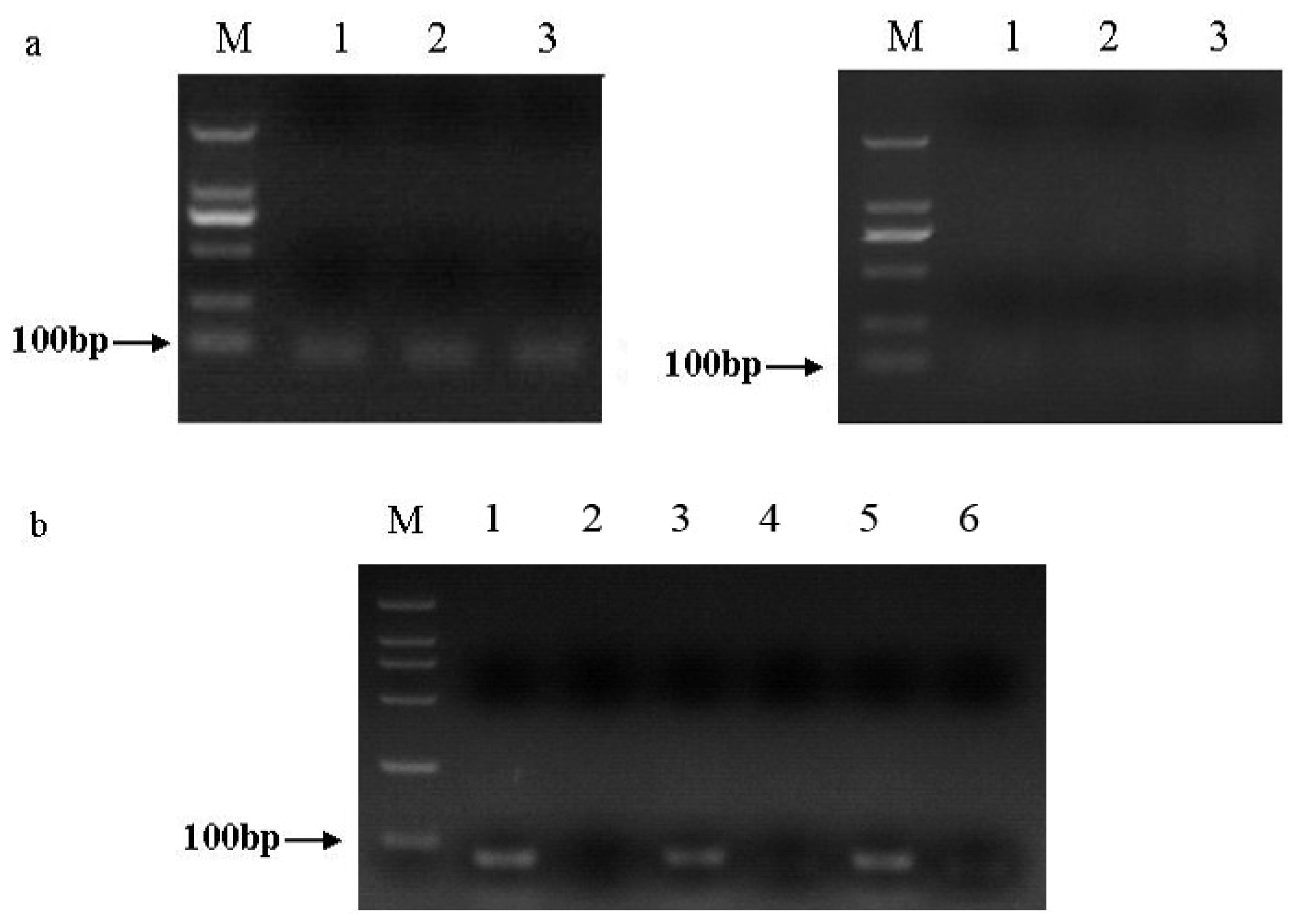
2.1. Amplification and Identification of sli-miR-14, sli-miR-2a and sli-bantam from S. litura
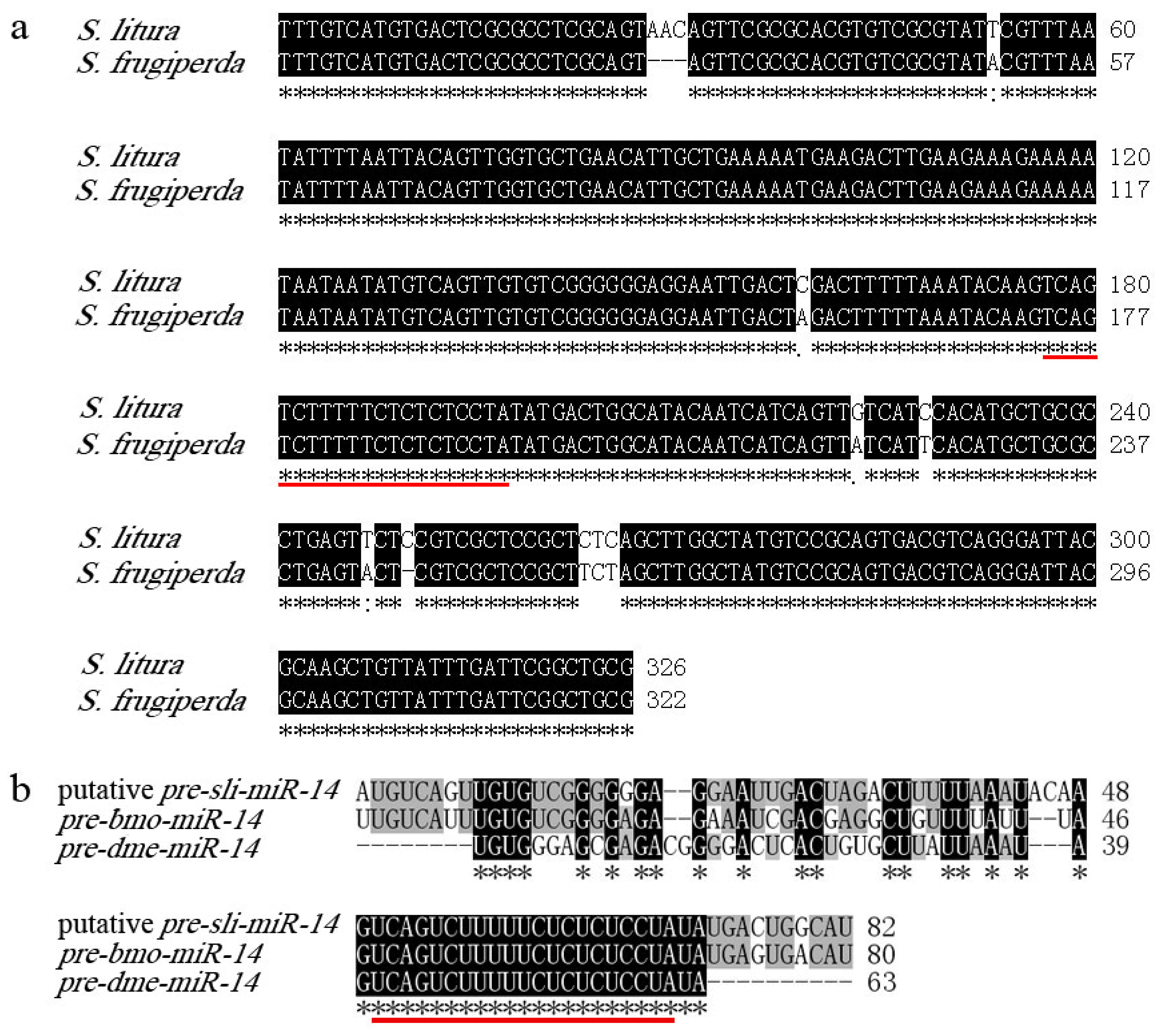
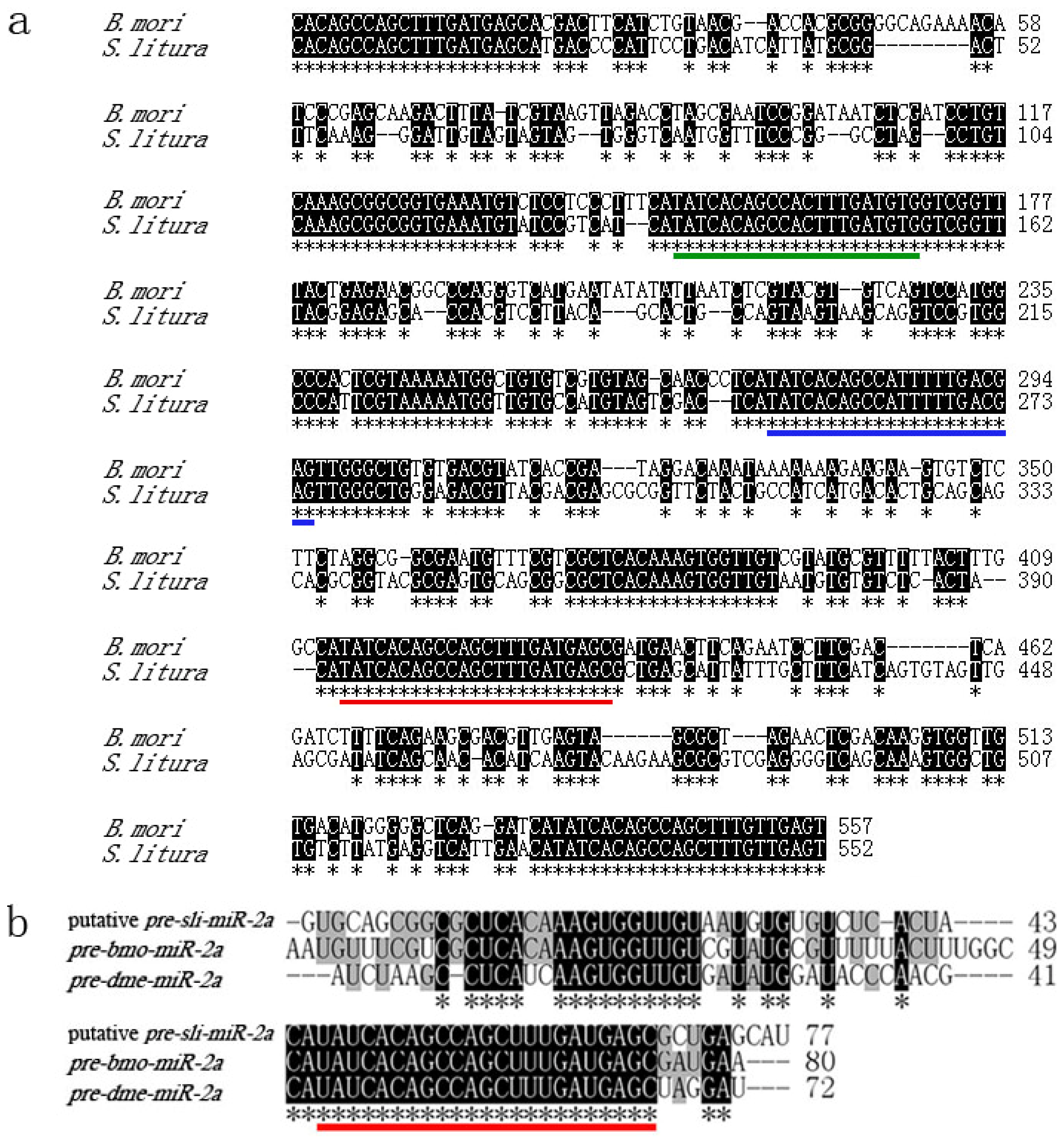
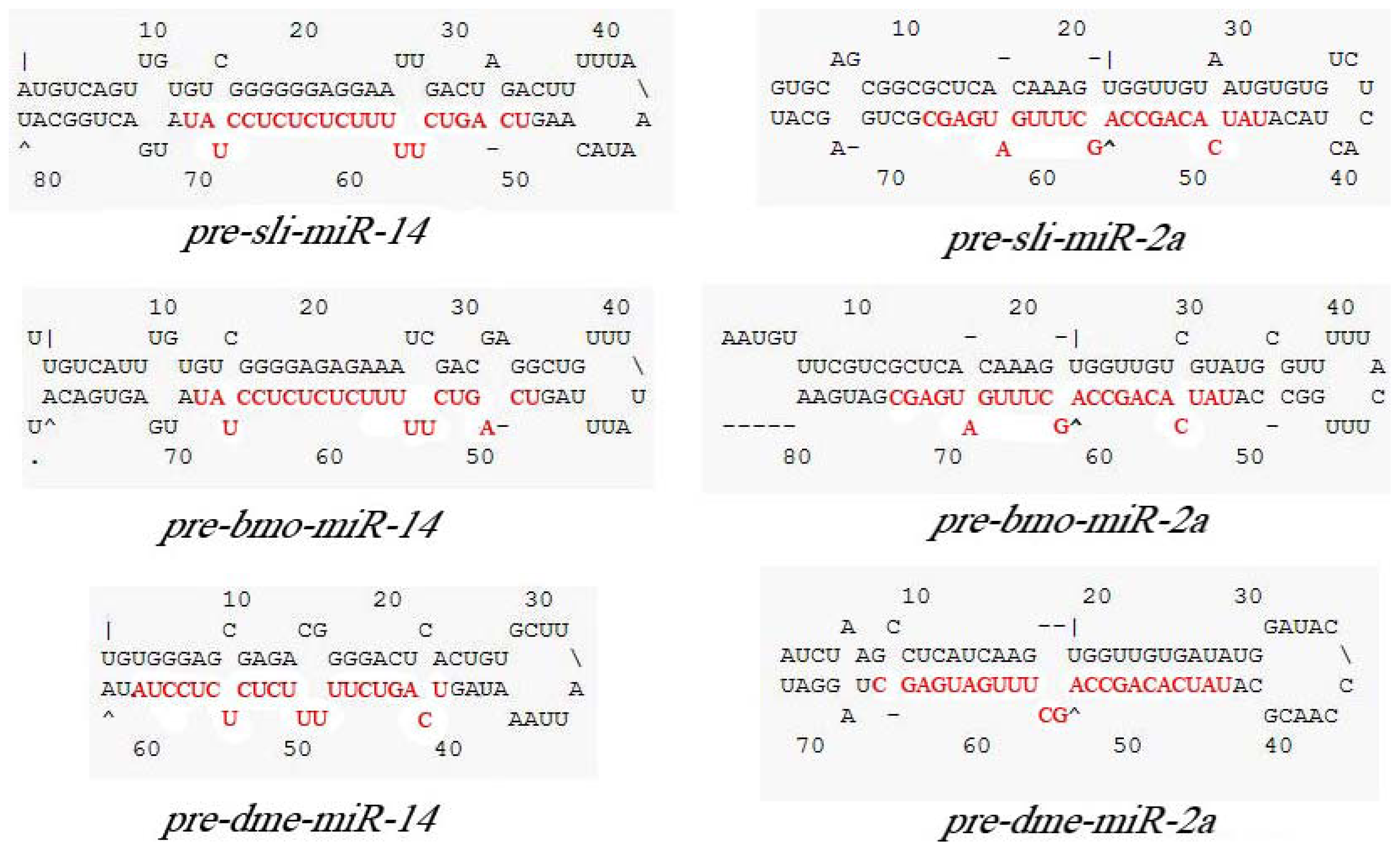
2.2. Cloning and Analysis of pre-miR-14 and pre-miR-2a from S. litura
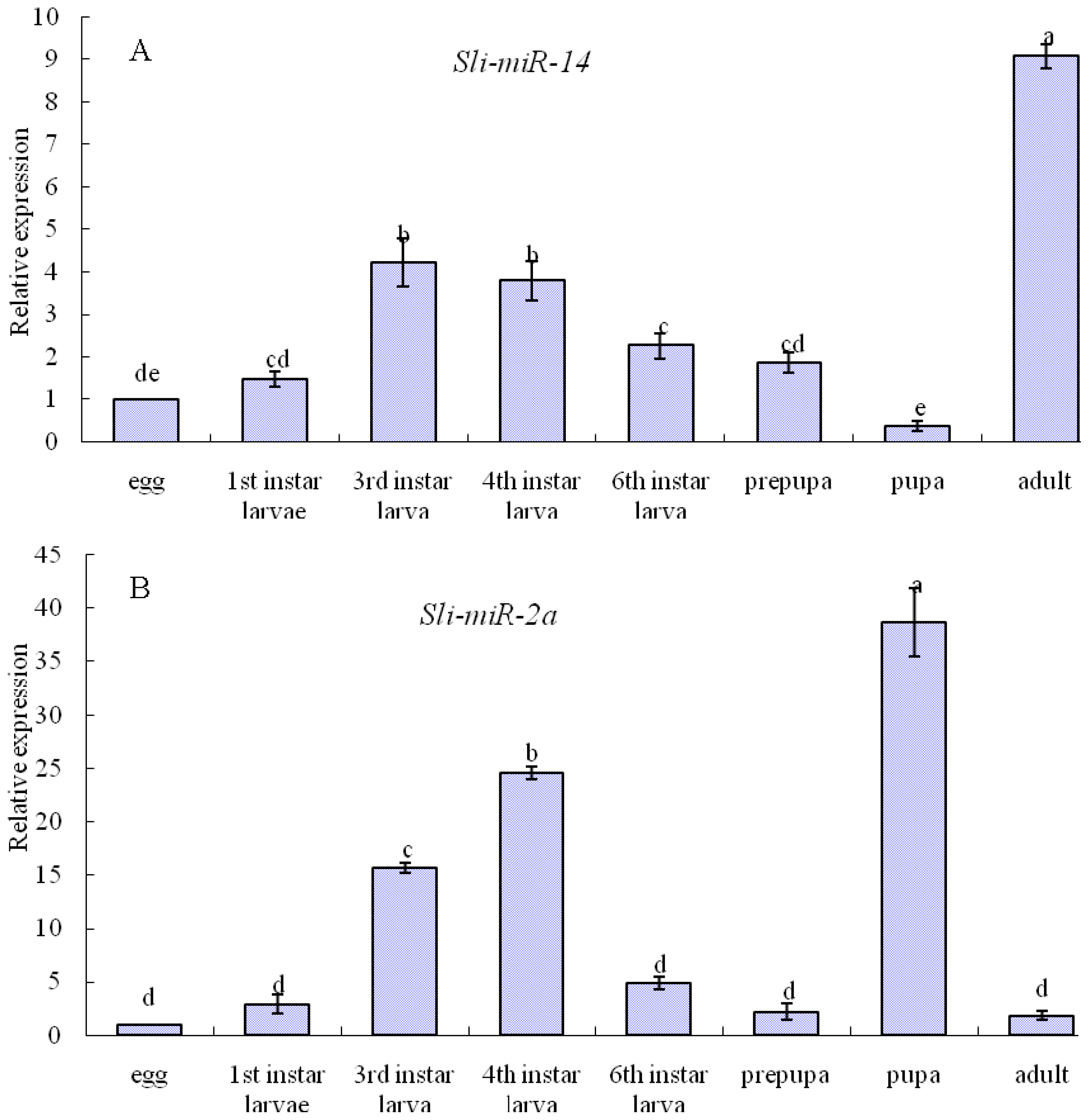
2.3. Expression Patterns of sli-miR-14, sli-miR-2a and sli-bantam from S. litura
2.4. Discussion
3. Experimental Section
3.1. Sample Preparation
3.2. Stem-Loop RT-PCR Primers Designing
3.3. RNA Extraction and Reverse Transcription
3.4. PCR of sli-miR-14, sli-miR-2a and sli-bantam
3.5. Pre-miRNA Primers Designing
3.6. Genomic DNA Isolation and the Amplification of pre-miR-14 and pre-miR-2a from S. litura
3.7. Real-Time Quantitative PCR
3.8. Sequence and Data Analysis
4. Conclusion
Acknowledgments
- Conflict of Interest The authors declare no conflict of interest.
References
- Valencia-Sanchez, M.A.; Liu, J.D.; Hannon, G.J.; Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 2006, 20, 515–524. [Google Scholar]
- Behura, S.K. Insect microRNAs: Structure, function and evolution. Insect Biochem. Mol. Biol 2007, 37, 3–9. [Google Scholar]
- Motameny, S.; Wolters, S.; Nürnberg, P.; Schumacher, B. Next generation sequencing of miRNAs-strategies, resources and methods. Genes 2010, 1, 70–84. [Google Scholar]
- Moulton, J.D.; Jiang, S. Gene knockdowns in adult animals: PPMOs and vivo-morpholinos. Molecules 2009, 14, 1304–1323. [Google Scholar]
- Chira, P.; Vareli, K.; Sainis, I.; Papandreou, C.; Briasoulis, E. Alterations of microRNAs in solid cancers and their prognostic value. Cancers 2010, 2, 1328–1353. [Google Scholar]
- Wienholds, E.; Plasterk, R.H.A. MicroRNA function in animal development. FEBS Lett 2005, 579, 5911–5922. [Google Scholar]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res 2008, 36, D154–D158. [Google Scholar]
- Kim, V.N.; Han, J.J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol 2009, 10, 126–139. [Google Scholar]
- Zhu, C.; Ji, C.B.; Zhang, C.M.; Gao, C.L.; Zhu, J.; Qin, D.N.; Kou, C.Z.; Zhu, G.Z.; Shi, C.M.; Guo, X.R. The lin-4 gene controls fat accumulation and longevity in Caenorhabditis elegans. Int. J. Mol. Sci 2010, 11, 4814–4825. [Google Scholar]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar]
- Xu, P.Z.; Vernooy, S.Y.; Guo, M.; Hay, B.A. The Drosophila microRNA miR-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol 2003, 13, 790–795. [Google Scholar]
- Garbuzov, A.; Tatar, M. Hormonal regulation of Drosophila microRNA let-7 and miR-125 that target innate immunity. Fly (Austin) 2010, 4, 306–311. [Google Scholar]
- Behura, S.K.; Whitfield, C.W. Correlated expression patterns of microRNA genes with age-dependent behavioural changes in honeybee. Insect Mol. Biol 2010, 19, 431–439. [Google Scholar]
- Yu, X.M.; Zhou, Q.; Li, S.C.; Luo, Q.B.; Cai, Y.M.; Lin, W.C.; Chen, H.; Yang, Y.; Hu, S.N.; Yu, J. The Silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages. PLoS One 2008, 3. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, X.; Ge, X.; Jiang, J.; Li, M.; Jia, S.; Yang, X.; Kan, Y.; Miao, X.; Zhao, G.; Li, F.; Huang, Y. Insect-specific microRNA involved in the development of the silkworm. Bombyx mori. PLoS One 2009, 4. [Google Scholar] [CrossRef]
- Marco, A.; Hui, J.H.L.; Ronshaugen, M.; Griffiths-Jones, S. Functional shifts in insect microRNA evolution. Genome Biol. Evol 2010, 2, 686–696. [Google Scholar]
- Mead, E.A.; Tu, Z.J. Cloning, characterization, and expression of microRNAs from the Asian malaria mosquito. Anopheles stephensi. BMC Genomics 2008, 9. [Google Scholar] [CrossRef]
- Cao, J.; Tong, C.Z.; Wu, X.J.; Lv, J.N.; Yang, Z.L.; Jin, Y.F. Identification of conserved microRNAs in Bombyx mori (silkworm) and regulation of fibroin L chain production by microRNAs in heterologous system. Insect Biochem. Mol. Biol 2008, 38, 1066–1071. [Google Scholar]
- Fu, H.J.; Zhu, J.; Yang, M.; Zhang, Z.Y.; Tie, Y.; Jiang, H.; Sun, Z.X.; Zheng, X.F. A novel method to monitor the expression of microRNAs. Mol. Biotechnol 2006, 32, 197–204. [Google Scholar]
- Chen, C.F.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.H.; Lee, D.H.; Nguyen, J.Y.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; Lao, K.Q.; Livak, K.J.; Guegler, K.J. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005, 33, 179–187. [Google Scholar]
- Tang, F.C.; Hajkova, P.; Barton, S.C.; Lao, K.Q.; Surani, M.A. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res 2006, 34, 9–15. [Google Scholar]
- Smibert, P.; Lai, E.C. A view from Drosophila: Multiple biological functions for individual microRNAs. Semin. Cell Dev. Biol 2010, 21, 745–753. [Google Scholar]
- He, P.A.; Nie, Z.M.; Chen, J.Q.; Chen, J.; Lv, Z.B.; Sheng, Q.; Zhou, S.P.; Gao, X.L.; Kong, L.Y.; Wu, X.F.; Jin, Y.F.; Zhang, Y.Z. Identification and characteristics of microRNAs from. Bombyx mori. BMC Genomics 2008, 9. [Google Scholar] [CrossRef]
- Jishy, V.; Stephen, M.C. microRNA miR-14 acts to modulate a positive autoregulatory loop Drosophila controlling steroid hormone signaling in Drosophila. Genes Dev 2007, 21, 2277–2282. [Google Scholar]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the pro-apoptotic gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar]
- Stark, A.; Brennecke, J.; Russell, R.B.; Cohen, S.M. Identification of Drosophila microRNA targets. PLoS Biol 2003, 1. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Jiang, J.M.; Liu, Q.; Yang, L.Q. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 2004, 32. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−Δ ΔCt method. Methods 2001, 25, 402–408. [Google Scholar]
- Aravin, A.A.; Lagos-Quintana, M.; Yalcin, A.; Zavolan, M.; Marks, D.; Snyde, B.; Gaasterland, T.; Meyer, J.; Tuschl, T. The small RNA profile during Drosophila melanogaster development. Dev. Cell 2003, 5, 337–350. [Google Scholar]
- Huang, G.L.; Zhang, X.H.; Guo, G.L.; Huang, K.T.; Yang, K.Y.; Shen, X.; You, J.; Hu, X.Q. Clinical significance of miR-21 expression in breast cancer: SYBR-Green I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol. Rep 2009, 21, 673–679. [Google Scholar]

| RNA | linear equation | PCR efficiency | R2 |
|---|---|---|---|
| Sli-miR-14 | y = −3.3125x + 39.562 | 2.0039 | 0.9987 |
| Sli-miR-2a | y = −3.2811x + 36.678 | 2.0173 | 0.9995 |
| 5S | y = −3.3915x + 33.219 | 1.9718 | 0.9945 |
| miRNAs | Primer | Sequence (5′-3′) |
|---|---|---|
| Sli-miR-14 | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTAGGAG |
| Forward | CGGGGCTCAGTCTTTTTCT | |
| Reverse | GTGCAGGGTCCGAGGT | |
| Sli-miR-2a | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTCAT |
| Forward | GCCAGTATCACAGCCAGCT | |
| Reverse | GTGCAGGGTCCGAGGT | |
| Sli-bantam | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAATCAG |
| Forward | GCCCCGTGAGATCATTTTG | |
| Reverse | GTGCAGGGTCCGAGGT |
| miRNAs | Primer | Sequence (5′-3′) |
|---|---|---|
| Sli-miR-14 | RT | GCACTTCAGTGTCGTGGTCAGTGACGGCAATTTGAAGTGCTAGGAG |
| Forward | CGCACGACGCATCAGTCAGTCTTTTTCT | |
| Reverse | GCACTTCAGTGTCGTGGTCAGTGACGGCAATT | |
| Sli-miR-2a | RT | GCACTTCAGTGTCGTGGTCAGTGACGGCAATTTGAAGTGCGCTCAT |
| Forward | CGACACACACCATCAGTATCACAGCCAGCT | |
| Reverse | GCACTTCAGTGTCGTGGTCAGTGACGGCAATT | |
| Sli-bantam | RT | GCACTTCAGTGTCGTGGTCAGTGACGGCAATTTGAAGTGCAATCAG |
| Forward | CGCATCGTAGCATCGCTGAGATCATTTTG | |
| Reverse | GCACTTCAGTGTCGTGGTCAGTGACGGCAATT |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gao, L.; Zuo, H.; Liu, K.; Li, H.; Zhong, G. A New Strategy for Identification of Highly Conserved microRNAs in Non-Model Insect, Spodoptera litura. Int. J. Mol. Sci. 2012, 13, 612-627. https://doi.org/10.3390/ijms13010612
Gao L, Zuo H, Liu K, Li H, Zhong G. A New Strategy for Identification of Highly Conserved microRNAs in Non-Model Insect, Spodoptera litura. International Journal of Molecular Sciences. 2012; 13(1):612-627. https://doi.org/10.3390/ijms13010612
Chicago/Turabian StyleGao, Lu, Hongliang Zuo, Keling Liu, Haiyi Li, and Guohua Zhong. 2012. "A New Strategy for Identification of Highly Conserved microRNAs in Non-Model Insect, Spodoptera litura" International Journal of Molecular Sciences 13, no. 1: 612-627. https://doi.org/10.3390/ijms13010612




