A Chitosan Induced 9-Lipoxygenase in Adelostemma gracillimum Seedlings
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of a Lipoxygenase cDNA from Seedlings of Adelostemma gracillimum
2.2. Comparison between AgLOX1 and Other Plant Lipoxygenase Genes
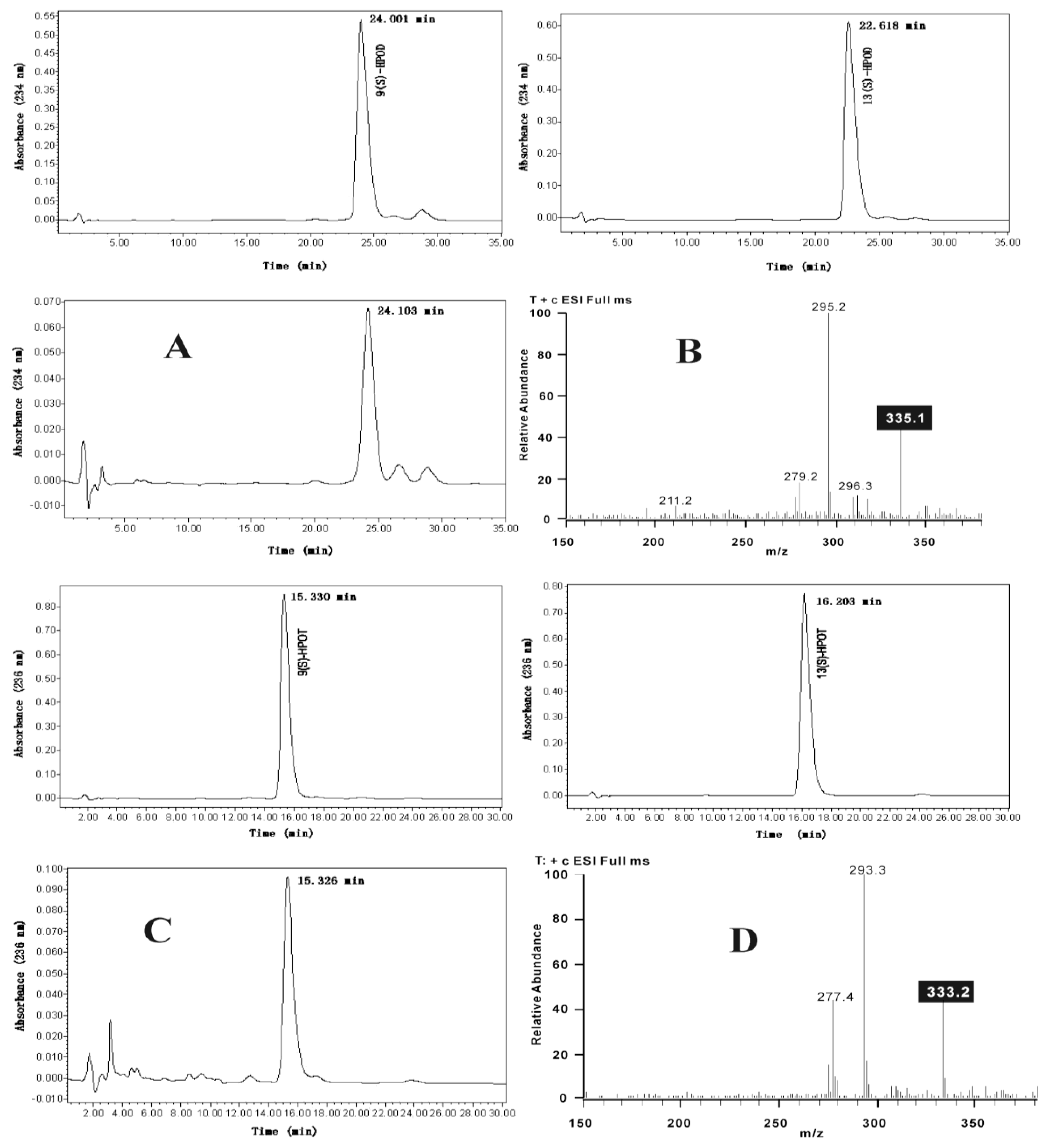
2.3. Functional Analysis of AgLOX1
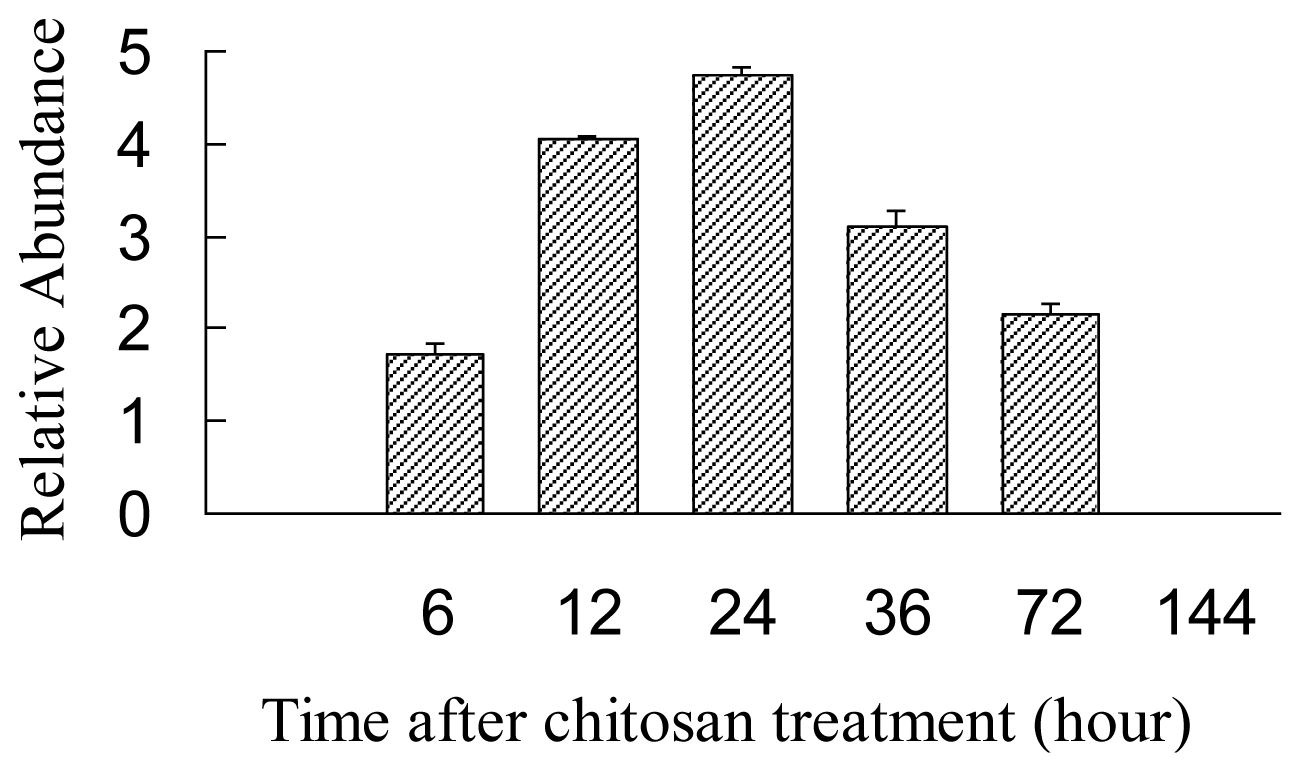
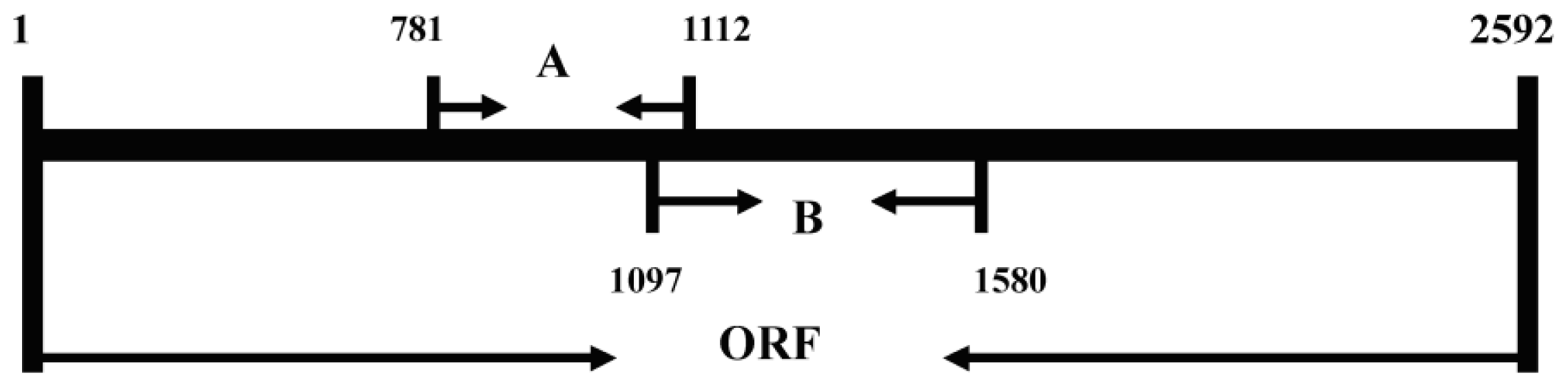
2.4. Expression Analysis of AgLOX1 in Seedling upon Chitosan Treatment
3. Experimental Section
3.1. Plant Materials, Substrates and Reagents
3.2. Extraction and LC-ESI-MS Analysis of Trihydroxy Octadecenoic Acids
3.3. cDNA Synthesis and RACE
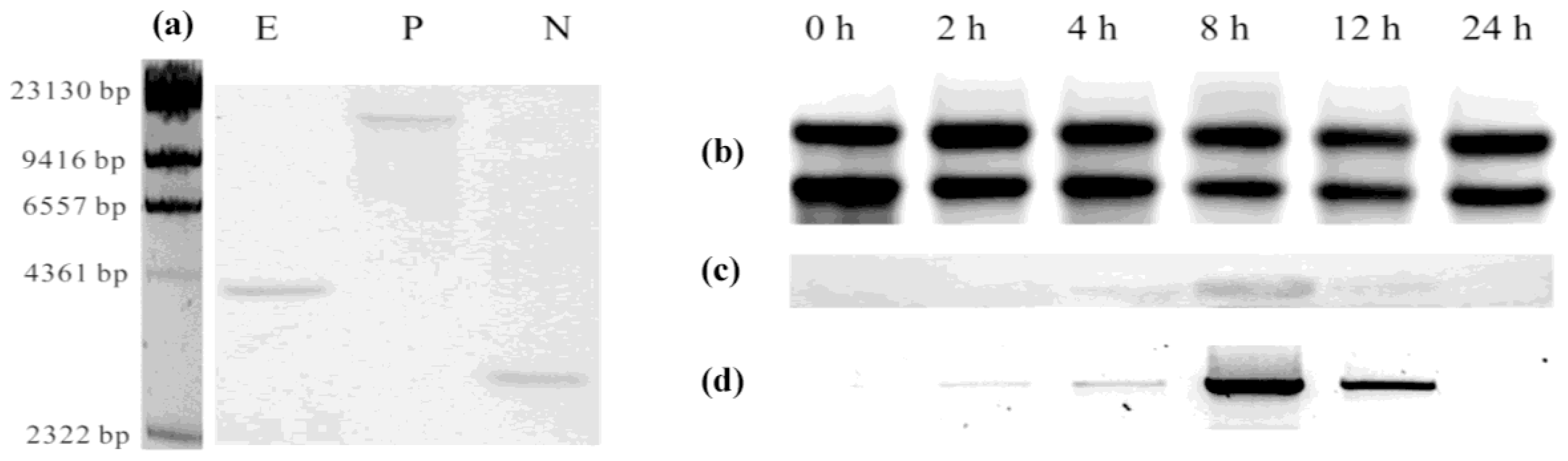
3.4. Southern and Northern Analyses
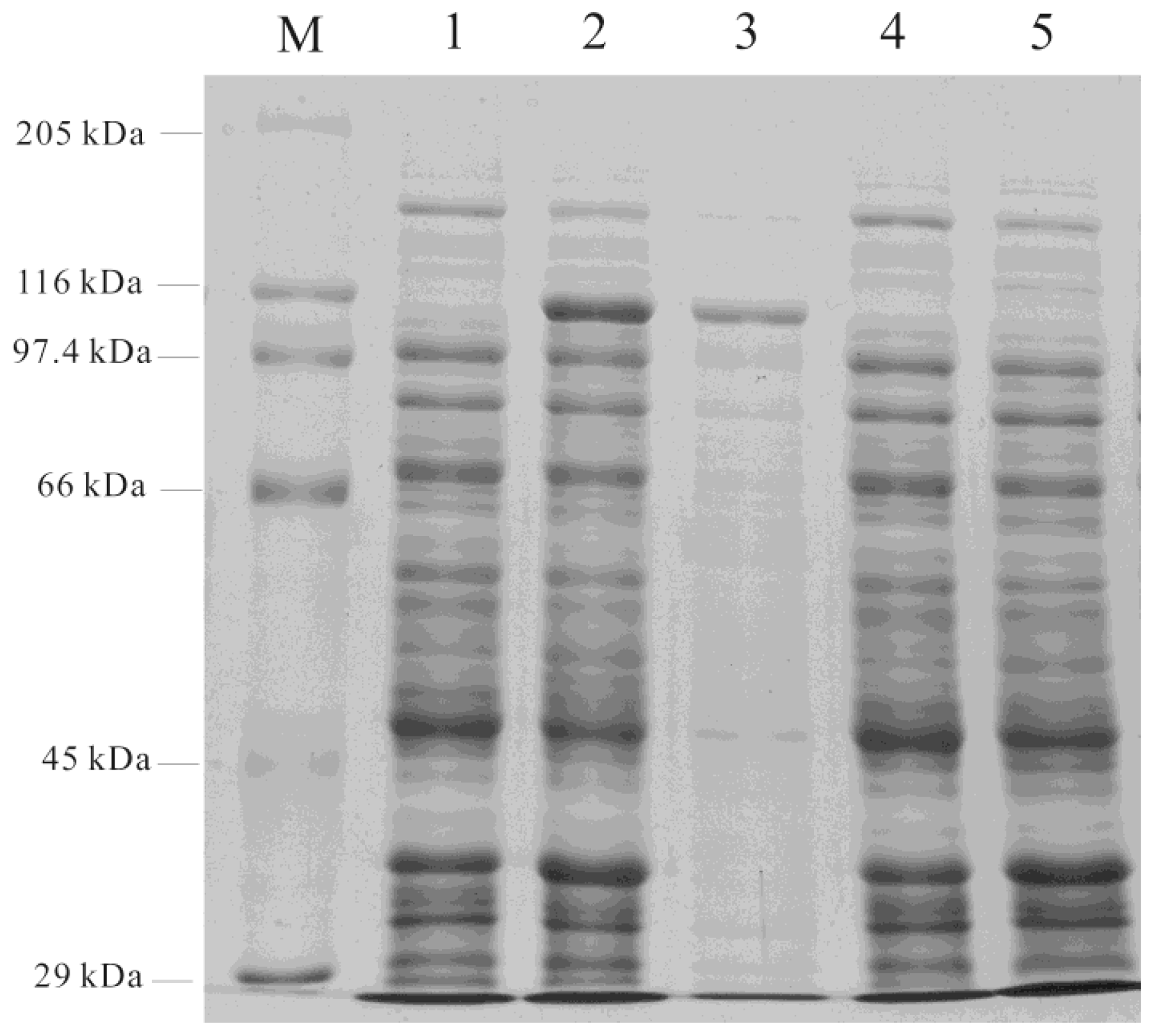
3.5. Expression of cDNA in E. coli
3.6. Enzyme Assays and Products Analysis
4. Conclusions
Acknowledgements
References
- Siedow, J.N. Plant lipoxygenase: Structure and function. Annu. Rev. Plant Physiol. Plant Mol. Biol 1991, 42, 145–188. [Google Scholar]
- Mosblech, A.; Feussner, I.; Heilmann, I. Oxylipins: Structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem 2009, 47, 511–517. [Google Scholar]
- Gardner, H.W. Recent investigations into the lipoxygenase pathway of plants. Biochim. Biophys. Acta 1991, 1084, 221–239. [Google Scholar]
- Brash, A.R. Lipoxygenases: Occurrence, functions, catalysis, and acquisition of Substrate. J. Biol. Chem 1999, 274, 23679–23682. [Google Scholar]
- Feussner, I.; Kühn, H.; Wasternack, C. Lipoxygenase-dependent degradation of storage lipids. Trends Plant Sci 2001, 6, 268–273. [Google Scholar]
- Bhardwaj, P.K.; Kaur, J.; Sobti, R.C.; Ahuja, P.S.; Kumar, S. Lipoxygenase in Caragana jubata responds to low temperature, abscisic acid, methyl jasmonate and salicylic acid. Gene 2011, 483, 49–53. [Google Scholar]
- Melan, M.A.; Dong, X.N.; Endara, M.E.; Davis, K.R.; Ausubel, F.M.; Peterman, T.K. An Arabidopsis thaliana lipoxygenase gene can be induced by pathogens, abscisic acid, and methy jasmonate. Plant Physiol 1993, 101, 441–450. [Google Scholar]
- Blée, E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci 2002, 7, 315–321. [Google Scholar]
- Grechkin, A. Recent developments in biochemistry of the plant lipoxygenase pathway. Prog. Lipid Res 1998, 37, 317–352. [Google Scholar]
- Göbel, C.; Feussner, I.; Schmidt, A.; Scheel, D.; Sanchez-Serrano, J.; Hamberg, M.; Rosahl, S. Oxylipin profiling reveals the preferential stimulation of the 9-lipoxygenase pathway in elicitor-treated potato cells. J. Biol. Chem 2001, 276, 6267–6273. [Google Scholar]
- Montillet, J.L.; Agnel, J.P.; Ponchet, M.; Vailleau, F.; Roby, D.; Triantaphylidès, C. Lipoxygenase-mediated production of fatty acid hydroperoxides is a specific signature of the hypersensitive reaction in plants. Plant Physiol. Biochem 2002, 40, 633–639. [Google Scholar]
- Zhao, P.J.; Gan, F.Y.; Shen, Y.M. Tissue Culture of Adelostemma gracillimum and Multihydroxy Fatty Acid in Calli. Proceedings of the 70th Anniversary of the Botanical Society of China, Lanzhou, China, 4–8 July 2003; High Education Press: Beijing, China, 2003; pp. 441–442. [Google Scholar]
- Spiteller, G. Lipid peroxidation in aging and age-dependent diseases. Exp. Gerontol 2001, 36, 1425–1457. [Google Scholar]
- Kolomiets, M.V.; Hannapel, D.J.; Gladon, R.J. Nucleotide sequence of a cDNA clone for a lipoxygenase from abscisic acid-treated potato leaves. Plant Physiol 1996, 112, 445. [Google Scholar]
- Véronési, C.; Fournier, J.; Rickauer, M.; Marolda, M.; Esquerré-Tugayé, M.T. Nucleotide sequence of an elicitor-induced tobacco lipoxygenase cDNA (GenBank X84040). Plant Physiol 1995, 108, 1342. [Google Scholar]
- Boyington, J.C.; Gaffney, B.J.; Amzel, L.M. The three dimentional structure of an arachidonic acid 15-lipoxygenase. Science 1993, 260, 1482–1486. [Google Scholar]
- Casey, R. Lipoxygenases. In Seed Proteins; Shewry, P.R., Casey, R., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1999; pp. 865–708. [Google Scholar]
- Shibata, D.; Slusarenko, A.J.; Casey, R.; Hildebrand, D.; Bell, E. Lipoxygenases. Plant Mol. Biol. Rep 1994, 12, S41–S42. [Google Scholar]
- Hornung, E.; Walther, M.; Kuhn, H.; Feussner, I. Conversion of cucumber linoleate 13-lipoxygenase to a 9-lipoxygenase species by side-directed mutagenesis. Proc. Natl. Acad. Sci. USA 1999, 96, 4192–4197. [Google Scholar]
- Ma, C.L.; Li, J.; Zhao, P.J. Tissue culture of Adelostemma gracillimum seedlings and the inducement of lipoxygenase. Guihaia 2009, 29, 386–389. [Google Scholar]
- Komaraiah, P.; Amrutha, R.A.; Kisho, P.B.K.; Ramakrishna, S.V. Elicitor enhanced production of plumbagin in suspension cultures of Plumbago rosea L. Enzym. Microb. Technol 2002, 31, 634–639. [Google Scholar]
- Farmer, E.E.; Ryan, C.A. Interplant communication. Airbone methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl. Acad. Sci. USA 1990, 87, 7713–7716. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar]
- Doares, S.H.; Syrovets, T.; Weiler, E.W.; Ryan, C.A. Oligogalacturonides and chitosan activate plant defensive genes through the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 1995, 92, 4095–4098. [Google Scholar]
- Matsui, K.; Minami, A.; Hornung, E.; Shibata, H.; Kishimoto, K.; Ahnert, V.; Kindl, H.; Kajiwara, T.; Feussner, I. Biosynthesis of fatty acid derived aldehydes is induced upon mechanical wounding and its products show fungicidal activities in cucumber. Phytochemistry 2006, 67, 649–657. [Google Scholar]
- Hamberg, M. An epoxy alcohol synthase pathway in higher plants: Biosynthesis of antifungal trihydroxy oxylipins in leaves of potato. Lipids 1999, 34, 1131–1142. [Google Scholar]
- Xu, Q.M.; Liu, Y.L.; Li, X.R.; Li, X.; Yang, S.L. Three new fatty acids from the roots of Boehmeria nivea (L.) Gaudich and their antifungal activities. 2011, 25, 640–647. [Google Scholar]
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, J.; Zhao, P.-J.; Ma, C.-L.; Zeng, Y. A Chitosan Induced 9-Lipoxygenase in Adelostemma gracillimum Seedlings. Int. J. Mol. Sci. 2012, 13, 540-551. https://doi.org/10.3390/ijms13010540
Li J, Zhao P-J, Ma C-L, Zeng Y. A Chitosan Induced 9-Lipoxygenase in Adelostemma gracillimum Seedlings. International Journal of Molecular Sciences. 2012; 13(1):540-551. https://doi.org/10.3390/ijms13010540
Chicago/Turabian StyleLi, Jing, Pei-Ji Zhao, Chang-Le Ma, and Ying Zeng. 2012. "A Chitosan Induced 9-Lipoxygenase in Adelostemma gracillimum Seedlings" International Journal of Molecular Sciences 13, no. 1: 540-551. https://doi.org/10.3390/ijms13010540



