Genetic Characterization of Five Hatchery Populations of the Pacific Abalone (Haliotis discus hannai) Using Microsatellite Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Microsatellite Genotyping
2.3. Data Analysis
3. Results
3.1. Genetic Variability
3.2. Genetic Differentiation among Populations
4. Discussion
5. Conclusions
Acknowledgments
References
- Elliot, NG. Genetic improvement programs in abalone: what is the future? Aquacult. Res 2000, 31, 51–59. [Google Scholar]
- Zhang, G; Wang, Z; Chang, Y; Song, J; Ding, J; Wang, Y; Wang, R. Triploid induction in Pacific abalone Haliotis discus hannai ino by 6–dimethylaminopurine and the performance of triploid juveniles. J. Shellfish Res 1998, 17, 783–788. [Google Scholar]
- Shin, YT; Jang, HS; Kim, BT. Production Control and Distribution Reform as a Way to Develop the Abalone Aquaculture Industry; Korea Mariculture Institute Press, Korea Mariculture Institute: Seoul, Korean, 2008. [Google Scholar]
- MIFAFF–Ministry for Food, Agriculture, Forestry and Fisheries of Korea. Annual Statistics of Fisheries Production in 2009; Ministry for Food, Agriculture, Forestry and Fisheries: Seoul, Korean, 2009. [Google Scholar]
- MOMAFK–Ministry of Maritime Affairs and Fisheries of Korea. Annual Statistics of Fisheries Production in 2007; Ministry of Maritime Affairs and Fisheries: Seoul, Korean, 2007. [Google Scholar]
- Li, Q; Park, C; Endo, T; Kijima, A. Loss of genetic variation at microsatellite loci in hatchery strains of the Pacific abalone (Haliotis discus hannai). Aquaculture 2004, 235, 207–222. [Google Scholar]
- Tauz, D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucl. Acids Res 1989, 17, 6463–6471. [Google Scholar]
- Holland, BS. Invasion without a bottleneck: Microsatellite variation in natural and invasive populations of the brown mussel Perna perna (L). Mar. Biotechnol 2001, 3, 407–415. [Google Scholar]
- Cheng, Y; Xinping, Z; Xiaowen, S. Development of microsatellite markers and their utilization in genetic diversity analysis of cultivated and wild populations of the mud carp (Cirrhina molitorella). J. Genet. Genomics 2008, 35, 201–206. [Google Scholar]
- Lemay, MA; Boulding, EG. Microsatellite pedigree analysis reveals high variance in reproductive success and reduced genetic diversity in hatchery-spawned northern abalone. Aquaculture 2009, 295, 22–29. [Google Scholar]
- Liu, F; Xia, JH; Bai, ZY; Fu, JJ; Li, JL; Yue, GH. High genetic diversity and substantial population differentiation in grass carp (Ctenopharyngodon idella) revealed by microsatellite analysis. Aquaculture 2009, 297, 51–56. [Google Scholar]
- Marchant, S; Haye, PA; Marin, SA; Winkler, FM. Genetic variability revealed with microsatellite markers in an introduced population of the abalone Haliotis discus hannai Ino. Aquacult. Res 2009, 40, 298–304. [Google Scholar]
- Yue, GH; Zhu, ZY; Lo, LC; Wang, CM; Lin, G; Fenf, F; Pang, HY; Li, J; Gong, P; Liu, HM; Tan, J; Chou, R; Lim, H; Orban, L. Genetic variation and population structure of Asian seabass (Lates calcarifer) in the Asia-Pacific region. Aquaculture 2009, 293, 22–28. [Google Scholar]
- Sekino, M; Hara, M. Microsatellite loci in Pacific abalone, Haliotis discus discus (Mollusca, Gastropoda, Haliotidae). Mol. Ecol. Notes 2001, 1, 8–10. [Google Scholar]
- Li, Q; Park, C; Kijima, A. Isolation and characterization of microsatellite loci in the Pacific abalone, Haliotis discus hannai. J. Shellfish Res 2002, 21, 811–815. [Google Scholar]
- Sekino, M; Saido, T; Fujita, T; Kobayashi, T; Takami, H. Microsatellite DNA markers of Ezo abalone (Haliotis discus hannai): A preliminary assessment of natural populations sampled from heavily stocked areas. Aquaculture 2005, 243, 33–47. [Google Scholar]
- An, HS; Hong, SW; Kim, EM; Lee, JH; Noh, JK; Kim, HC; Park, CJ; Min, BH; Myeong, JI. Comparative genetic diversity of wild and released populations of Pacific abalone Haliotis discus discus in Jeju, Korea, based on cross-species microsatellite markers including two novel loci. Anim. Cells Syst 2010, 14, 305–313. [Google Scholar]
- An, HS; Lee, JH; Dong, CM; Noh, JK; Kim, HC; Park, CJ; Park, KD; Min, BH; Park, JW; Myeong, JI. New polymorphic microsatellite markers in Pacific abalone Haliotis discus hannai and their application to genetic characterization of wild and aquaculture populations. Genes Genomics 2010, 32, 413–418. [Google Scholar]
- Hara, M; Sekino, M. Genetic differences between hatchery stocks and natural populations in Pacific Abalone (Haliotis discus) estimated using microsatellite DNA markers. Mar. Biotechnol 2007, 9, 74–81. [Google Scholar]
- Li, Q; Shu, J; Yu, R; Tian, C. Genetic variability of cultured populations of the Pacific abalone (Haliotis discus hannai Ino) in China based on microsatellites. Aquacult. Res 2007, 38, 981–990. [Google Scholar]
- van Oosterhout, C; Hutchinson, WF; Wills, DPM; Shipley, P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar]
- Beaumont, MA; Nichols, RA. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B 1996, 263, 1619–1626. [Google Scholar]
- Tiago, A; Ana, L; Ricardo, JL; Albano, B-P; Gordon, L. LOSITAN: A workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinforma 2008, 9, 323–327. [Google Scholar]
- Goudet, J. FSTAT: A computer program to calculate F-statistics. J. Hered 2002, 86, 485–486. [Google Scholar]
- El Mousadik, A; Petit, RJ. High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L.) Skeels) endemic to Morocco. Theor. Appl. Genet 1996, 92, 832–839. [Google Scholar]
- Excoffier, L; Smouse, PE; Quattro, JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar]
- Excoffier, L; Laval, G; Schneider, S. ARLEQUIN version 3.0: an integrated software package for population genetics data analysis. Evol. Bioinf 2005, 1, 47–50. [Google Scholar]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Weir, BS; Cockerham, CC. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Rousset, F. Equilibrium values of measures of population subdivision for stepwise mutation processes. Genetics 1996, 142, 1357–1362. [Google Scholar]
- Rice, WR. Analyzing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar]
- Cavalli-Sforza, LL; Edwards, AWF. Phylogenetic analysis: models and estimation procedures. Evolution 1967, 32, 550–570. [Google Scholar]
- Takezaki, N; Nei, M. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 1996, 144, 389–399. [Google Scholar]
- Cornuet, JM; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar]
- Hara, M; Sekino, M. Genetic dufference between Ezo-awabi Haliotis discus hannai and Kuro-awabi H. discus discus populations: Microsatellite-based population analysis in Japanese abalone. Fish. Sci 2005, 71, 754–766. [Google Scholar]
- English, LJ; Maguire, GB; Ward, RD. Genetic variation of wild and hatchery populations of the pacific oyster, Crassostrea gigas (Thunberg) in Australia. Aquaculture 2000, 187, 283–298. [Google Scholar]
- Yu, H; Li, Q. Genetic variation of wild and hatchery populations of the Pacific oyster Crassostrea gigas assessed by microsatellite markers. J. Genet. Genomics 2007, 34, 1114–1122. [Google Scholar]
- Evans, B; Bartlett, J; Sweijd, N; Cook, P; Elliott, NG. Loss of genetic variation at microsatellite loci in hatchery produced abalone in Australia (Haliotis rubra) and South Africa (Haliotis midae). Aquaculture 2004, 233, 109–127. [Google Scholar]
- Zhang, Q; Allen, SK, Jr; Reece, KS. Genetic variation in wild and hatchery stocks of Suminoe oyster (Crassostrea ariakensis) assessed by PCR-RFLP and microsatellite markers. Mar. Biotechnol 2005, 7, 588–599. [Google Scholar]
- Addison, JA; Hart, MW. Spawning, copulation and inbreeding coefficients in marine invertebrates. Biol. Lett 2005, 1, 450–453. [Google Scholar]
- Estoup, A; Tailliez, C; Cornuet, J; Solignac, M. Size homoplasy and mutational processes of interrupted microsatellites in two bee spices, Apis mellifera and Bombus terrestris (Apidae). Mol. Biol. Evol 1995, 12, 1074–1084. [Google Scholar]
- Spencer, CC; Neigel, JE; Leberg, PL. Experimental evaluation of the usefulness of microsatellite DNA for detecting demographic bottlenecks. Mol. Ecol 2000, 9, 1517–1528. [Google Scholar]
- Launey, S; Hedgecock, D. High genetic load in the Pacific oyster Crassostrea gigas. Genetics 2001, 159, 255–265. [Google Scholar]
- Sugita, M; Fujio, Y. Effect of genotypes at the Aat-locus on the survival and growth rates in the cultured Oyster. Tohoku J. Agric. Res 1982, 33, 42–49. [Google Scholar]
- Sekino, M; Harab, N; Taniguchi, N. Loss of microsatellite and mitochondrial DNA variation in hatchery strains of Japanese flounder Paralichthys olivaceus. Aquaculture 2002, 213, 101–122. [Google Scholar]
- Was, A; Wenne, R. Genetic differentiation in hatchery and wild sea trout (Salmo trutta) in the southern Baltic at microsatellite loci. Aquaculture 2002, 204, 493–506. [Google Scholar]
- Alarcón, JA; Magoulas, A; Georgakopoulos, A; Zouros, T; Zouros, E; Alvarez, MC. Genetic comparison of wild and cultivated European populations of the gilthead sea bream (Sparus aurata). Aquaculture 2004, 230, 65–80. [Google Scholar]
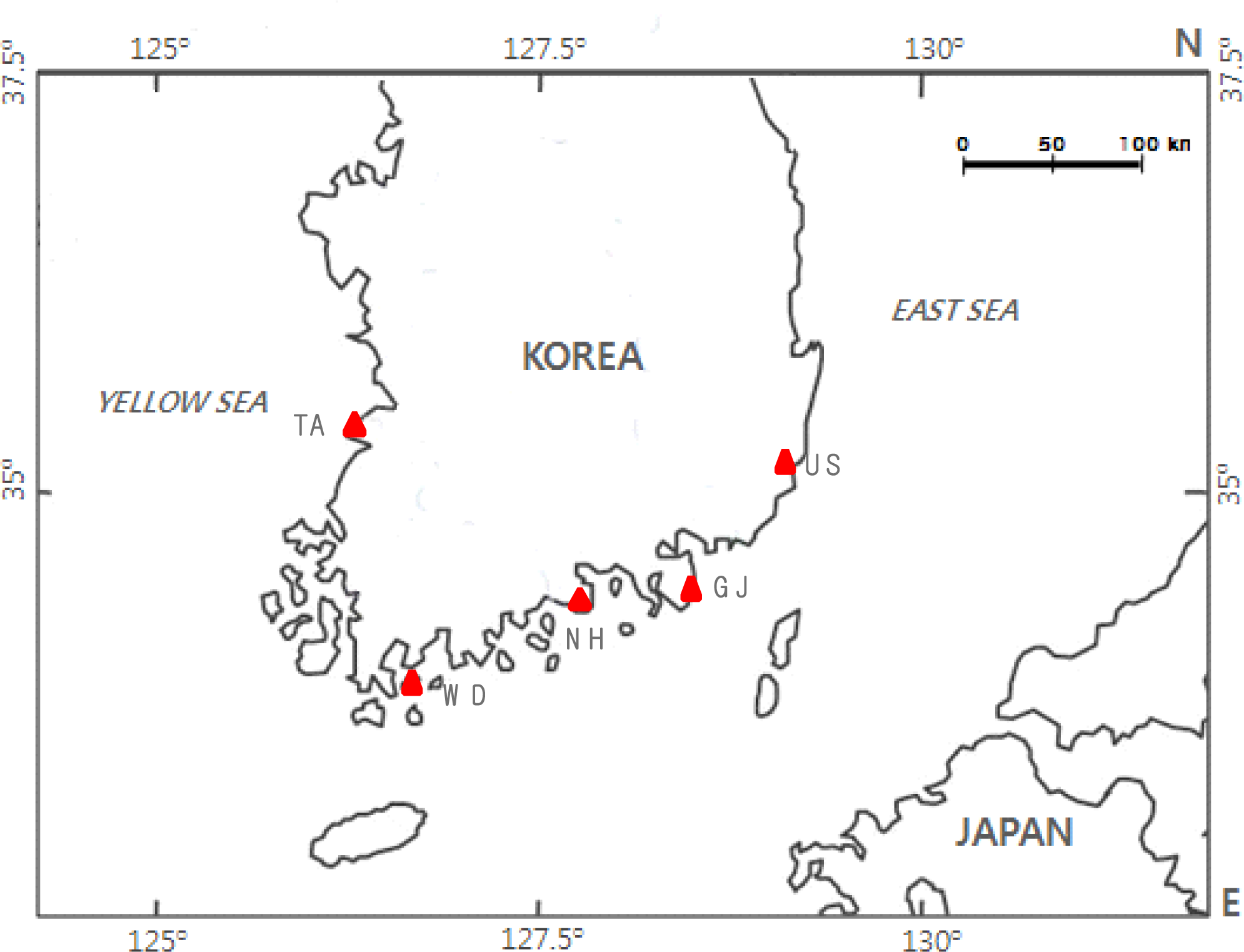


| Population’s Name (Abbreviation) | Sample Locality | Sample Size | Collection Date |
|---|---|---|---|
| Ulsan population (US) | Eastern Area (Ulsan); 35° 32′ N, 129° 25′ E | 51 | February. 2004 |
| Geoje population (GJ) | Southern Area (Geoje); 34° 53′ N, 128° 41′ E | 43 | June. 2003 |
| Namhae population (NH) | Southern Area (Namhae); 34° 42′ N, 128° 0′ E | 49 | June. 2003 |
| Wando population (WD) | Southern Area (Wando); 34° 23′ N, 126° 51′ E | 41 | May. 2003 |
| Taean population (TA) | Western Area (Taean); 36° 40′ N, 126° 16′ E | 39 | March. 2004 |
| Population (No.) | Microsatellite Loci | |||||||
|---|---|---|---|---|---|---|---|---|
| Hdh1321 | Hdd114B | Hdd229 | Hdh513 | Hdh512 | Hdh145 | Mean | ||
| Ulsan (51) | NA | 29 | 26 | 22 | 39 | 31 | 10 | 26.17 |
| AR | 27.59 | 24.07 | 20.40 | 34.45 | 28.70 | 9.23 | 24.07 | |
| S | 264–380 | 152–256 | 168–218 | 422.000 | 70–200 | 130–154 | ||
| U | 2 | 2 | 0 | 2 | 4 | 1 | 1.83 | |
| He | 0.961 | 0.943 | 0.913 | 0.967 | 0.964 | 0.750 | 0.916 | |
| Ho | 1.000 | 0.902 | 0.647 | 0.824 | 1.000 | 0.549 | 0.820 | |
| FIS | −0.041 | 0.044 | 0.293* | 0.149* | −0.037 | 0.270* | 0.106 | |
| P | 0.934 | 0.727 | 0.000* | 0.038 | 0.211 | 0.000* | ||
| Geoje (43) | NA | 29 | 23 | 21 | 31 | 23 | 10 | 22.83 |
| AR | 28.02 | 22.05 | 20.42 | 29.83 | 22.52 | 9.71 | 22.09 | |
| S | 264–366 | 152–252 | 172–226 | 190–422 | 88–148 | 114–154 | ||
| U | 0 | 1 | 2 | 3 | 2 | 1 | 1.50 | |
| He | 0.959 | 0.898 | 0.905 | 0.958 | 0.950 | 0.693 | 0.894 | |
| Ho | 0.814 | 0.930 | 0.674 | 0.744 | 0.953 | 0.488 | 0.767 | |
| FIS | 0.153 | −0.037 | 0.257* | 0.225 | −0.003 | 0.298* | 0.143 | |
| P | 0.000* | 0.940 | 0.000* | 0.011 | 0.001* | 0.000* | ||
| Namhae (49) | NA | 32 | 29 | 21 | 40 | 27 | 11 | 26.67 |
| AR | 28.92 | 27.04 | 20.16 | 36.38 | 25.36 | 10.83 | 24.78 | |
| S | 288–398 | 154–258 | 164–218 | 174–422 | 74–142 | 106–152 | ||
| U | 6 | 1 | 1 | 8 | 1 | 3 | 3.33 | |
| He | 0.925 | 0.951 | 0.945 | 0.968 | 0.950 | 0.777 | 0.919 | |
| Ho | 0.571 | 0.918 | 0.857 | 0.735 | 0.878 | 0.551 | 0.752 | |
| FIS | 0.385* | 0.034 | 0.094 | 0.243* | 0.077 | 0.293* | 0.184 | |
| P | 0.000* | 0.485 | 0.128 | 0.001* | 0.095 | 0.000* | ||
| Wando (41) | NA | 26 | 24 | 18 | 36 | 20 | 5 | 21.50 |
| AR | 25.60 | 23.46 | 17.56 | 35.16 | 19.89 | 5.00 | 21.11 | |
| S | 288–378 | 152–270 | 168–224 | 182–422 | 74–132 | 130–140 | ||
| U | 2 | 4 | 1 | 2 | 0 | 0 | 1.50 | |
| He | 0.945 | 0.930 | 0.879 | 0.969 | 0.934 | 0.651 | 0.885 | |
| Ho | 0.951 | 0.927 | 0.683 | 0.976 | 0.951 | 0.537 | 0.838 | |
| FIS | −0.007 | 0.004 | 0.225* | −0.007 | −0.019 | 0.178 | 0.054 | |
| P | 0.157 | 0.764 | 0.000* | 0.178 | 0.275 | 0.302 | ||
| Taean (39) | NA | 28 | 19 | 18 | 31 | 19 | 6 | 20.17 |
| AR | 28.00 | 19.00 | 18.00 | 31.00 | 19.00 | 6.00 | 20.17 | |
| S | 266–384 | 152–260 | 172–218 | 188–422 | 90–134 | 128–140 | ||
| U | 3 | 2 | 0 | 2 | 0 | 0 | 1.17 | |
| He | 0.945 | 0.907 | 0.925 | 0.964 | 0.922 | 0.781 | 0.907 | |
| Ho | 0.974 | 0.897 | 0.462 | 0.897 | 0.974 | 0.641 | 0.808 | |
| FIS | −0.031 | 0.010 | 0.505* | 0.070 | −0.058* | 0.181 | 0.111 | |
| P | 0.996 | 0.581 | 0.000* | 0.170 | 0.000* | 0.025 | ||
| Mean all populations | ||||||||
| NA | 28.80 | 24.20 | 20.00 | 35.40 | 24.00 | 8.40 | 23.47 | |
| AR | 27.63 | 23.12 | 19.31 | 33.36 | 23.10 | 8.15 | 22.44 | |
| U | 2.60 | 2.00 | 0.80 | 3.40 | 1.40 | 1.00 | 1.87 | |
| He | 0.951 | 0.926 | 0.909 | 0.965 | 0.944 | 0.730 | 0.904 | |
| Ho | 0.919 | 0.915 | 0.607 | 0.835 | 0.951 | 0.553 | 0.797 | |
| Population | Ulsan | Geoje | Namhae | Wando | Taean |
|---|---|---|---|---|---|
| Ulsan | – | 0.0068* (0.0744*) | 0.0050* (0.0341NS) | 0.0088* (0.0198NS) | 0.0091* (0.0053NS) |
| Geoje | 0.019 | – | 0.0115NS (0.0289*) | 0.0124* (0.0231NS) | 0.0176* (0.1016*) |
| Namhae | 0.018 | 0.024 | – | 0.0068* (−0.0082NS) | 0.0098* (0.0524*) |
| Wando | 0.019 | 0.021 | 0.020 | – | 0.0172* (0.0332*) |
| Taean | 0.018 | 0.026 | 0.023 | 0.020 | - |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
An, H.S.; Lee, J.W.; Kim, H.C.; Myeong, J.-I. Genetic Characterization of Five Hatchery Populations of the Pacific Abalone (Haliotis discus hannai) Using Microsatellite Markers. Int. J. Mol. Sci. 2011, 12, 4836-4849. https://doi.org/10.3390/ijms12084836
An HS, Lee JW, Kim HC, Myeong J-I. Genetic Characterization of Five Hatchery Populations of the Pacific Abalone (Haliotis discus hannai) Using Microsatellite Markers. International Journal of Molecular Sciences. 2011; 12(8):4836-4849. https://doi.org/10.3390/ijms12084836
Chicago/Turabian StyleAn, Hye Suck, Jang Wook Lee, Hyun Chul Kim, and Jeong-In Myeong. 2011. "Genetic Characterization of Five Hatchery Populations of the Pacific Abalone (Haliotis discus hannai) Using Microsatellite Markers" International Journal of Molecular Sciences 12, no. 8: 4836-4849. https://doi.org/10.3390/ijms12084836




