Novel Polymorphic Microsatellite Loci for the Korean Black Scraper (Thamnaconus modestus), and Their Application to the Genetic Characterization of Wild and Farmed Populations
Abstract
:1. Introduction
2. Results and Discussion
2.1. Microsatellite Loci Isolation
2.2. Genetic Characterization
2.3. Cross-Species Amplification
3. Experimental Section
3.1. Library Construction and Sequencing
3.2. Primer Design and Genotyping
3.3. Sample Comparisons
4. Conclusions
Acknowledgments
References
- Kim, PD. Miniature guide to whole Korean fishes. Scuba Diver 2007, 1, 215. [Google Scholar]
- Hauser, L; Adcock, GJ; Smith, PJ; Bernal Ramirez, J; Carvalho, GR. Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper. Proc. Natl. Acad. Sci. USA 2002, 99, 11742–11747. [Google Scholar]
- van Herwerden, L; Choat, JH; Dudgeon, CL; Carlos, G; Newman, SJ; Frisch, A; van Oppen, M. Contrasting patterns of genetic structure in two species of the coral trout Pletropomus (Serranidae) from east and west Australia: Introgressive hybridization or ancestral polymorphisms. Mol. Phylogenet. Evol 2006, 41, 420–435. [Google Scholar]
- Kim, MJ; An, HY; Choi, KH. Genetic characteristics of Pacific cod populations in Korea based on microsatellite markers. Fish. Sci 2010, 76, 595–603. [Google Scholar]
- Hamilton, MB; Pincus, EL; DiFiore, A; Fleischer, RC. Universal linker and ligation procedures for construction of genomic DNA libraries enriched for microsatellites. Biotechniques 1999, 27, 500–507. [Google Scholar]
- Gardner, MG; Cooper, SJB; Bull, CM; Grant, WN. Isolation of microsatellite loci from a social lizard, Egernia stokesii, using a modified enrichment procedure. J. Hered 1999, 90, 301–304. [Google Scholar]
- Carleton, KL; Streelman, JT; Lee, BY; Garnhart, N; Kidd, M; Kocher, TD. Rapid isolation of CA microsatellites from the tilapia genome. Anim. Genet 2002, 33, 140–144. [Google Scholar]
- An, HS; Park, JY; Kim, MJ; Lee, EY; Kim, KK. Isolation and characterization of microsatellite markers for the heavily exploited rockfish Sebastes schlegeli, and cross-species amplification in four related Sebastes spp. Conserv. Genet 2009, 10, 1969–1972. [Google Scholar]
- Sekino, M; Hara, M. Isolation and characterization of microsatellite DNA loci in Japanese flounder Paralichthys olivaceus (Pleuronectiformes, Pleuronectoidei, Paralichthyidae). Mol. Ecol 2000, 9, 2200–2203. [Google Scholar]
- Yokoyama, E; Sakamoto, T; Sugaya, T; Kitada, S. Six polymorphic microsatellite loci in the Japanese Spanish mackerel, Scomberomorus niphonius. Mol. Ecol. Notes 2006, 6, 323–324. [Google Scholar]
- DeWoody, JA; Avise, JC. Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J. Fish Biol 2000, 56, 461–473. [Google Scholar]
- Sekino, M; Hara, M; Taniguchi, N. Loss of microsatellite and mitochondrial DNA variation in hatchery strains of Japanese flounder Paralichthys olivaceus. Aquaculture 2002, 213, 101–122. [Google Scholar]
- Yue, GH; Li, Y; Chen, F; Lim, LC; Orban, L. Monitoring the genetic diversity of three Asian arowana (Scleropages formosus) captive stocks using AFLP and microsatellites. Aquaculture 2004, 237, 89–102. [Google Scholar]
- Callen, DF; Thompson, AD; Shen, Y; Phillips, HA; Mulley, JC; Sutherland, GR. Incidence and origin of “null” alleles in the (AC)n microsatellite markers. Am. J. Hum. Genet 1993, 52, 922–927. [Google Scholar]
- Excoffier, L; Laval, G; Schneider, S. ARLEQUIN version 3.0. An integrated software package for population genetics data analysis. Evol. Bioinf. Online 2005, 1, 47–50. [Google Scholar]
- Lynch, M; O’Hely, M. Captive breeding and the genetic fitness of natural populations. Conserv. Genet 2001, 2, 363–378. [Google Scholar]
- Hansen, MM. Estimating the long-term effects of stocking domesticated trout into wild brown trout (Salmo trutta) populations: an approach using microsatellite DNA analysis of historical and contemporary samples. Mol. Ecol 2002, 11, 1003–1015. [Google Scholar]
- Allendorf, FW; Phelps, SR. Loss of genetic variation in a hatchery stock of cutthroat trout. Trans. Am. Fish. Soc 1980, 109, 537–543. [Google Scholar]
- Vuorinen, J. Reduction of genetic variability in a hatchery stock of brown trout, Salmo trutta. J. Fish Biol 1984, 24, 339–348. [Google Scholar]
- Peakall, R; Gilmore, S; Keys, W; Morgante, M; Rafalski, A. Cross-species amplification of soybean (Glycine max) simple sequence repeats within the genus and other legume genera: Implications for the transferability of SSRs in plants. Mol. Biol. Evol 1998, 15, 1275–1287. [Google Scholar]
- Xu, Q; Liu, R. Development and characterization of microsatellite markers for genetic analysis of the swimming crab, Portunus trituberculatus. Biochem. Genet 2011, 49, 202–212. [Google Scholar]
- Asahida, T; Kobayashi, T; Saitoh, K; Nakayama, I. Tissue preservation and total DNA extraction from fish stored at ambient temperature using buffers containing high concentrations of urea. Fish Sci 1996, 62, 727–730. [Google Scholar]
- Li, Q; Park, C; Kijima, A. Isolation and characterization of microsatellite loci in the Pacific abalone, Haliotis discus hannai. J. Shellfish Res 2002, 212, 811–815. [Google Scholar]
- van Oosterhout, C; Hutchinson, WF; Wills, DPM; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar]
- Goudet, J. FSTAT version 2.9.3. A Program to Estimate and Test Gene Diversities and Fixation Indices. 2001. Available online: http://www2.unil.ch/popgen/softwares/fstat.htm accessed on 17 June 2011.
- Weir, BS; Cockerham, CC. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- El Mousadik, A; Petit, RJ. High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L.) Skeels) endemic to Morocco. Theor. Appl. Genet 1996, 92, 832–839. [Google Scholar]
- Slatkin, M; Excoffier, L. Testing for linkage disequilibrium in genotypic data using the EM algorithm. Heredity 1996, 76, 377–383. [Google Scholar]
- Rice, WR. Analyzing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar]
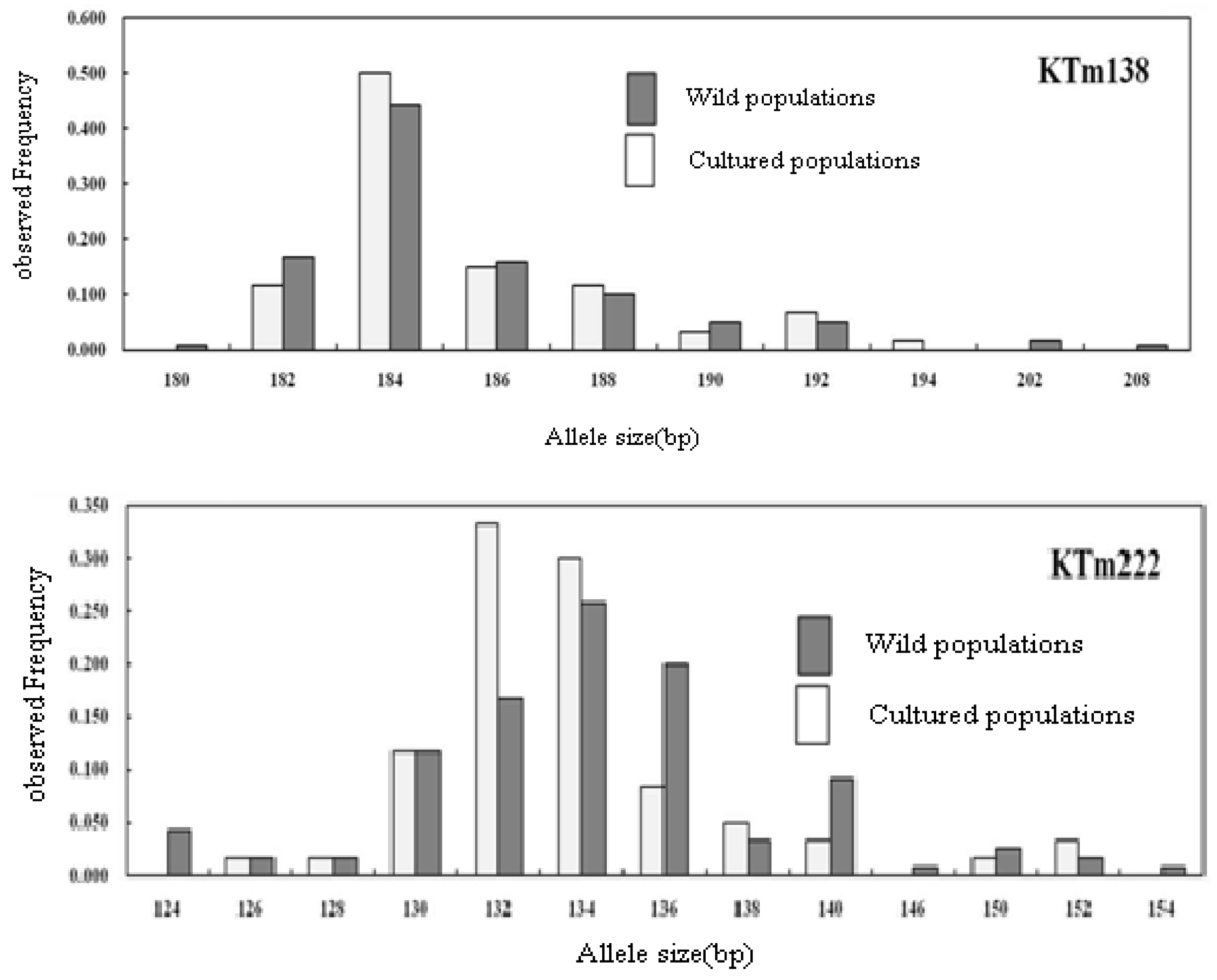

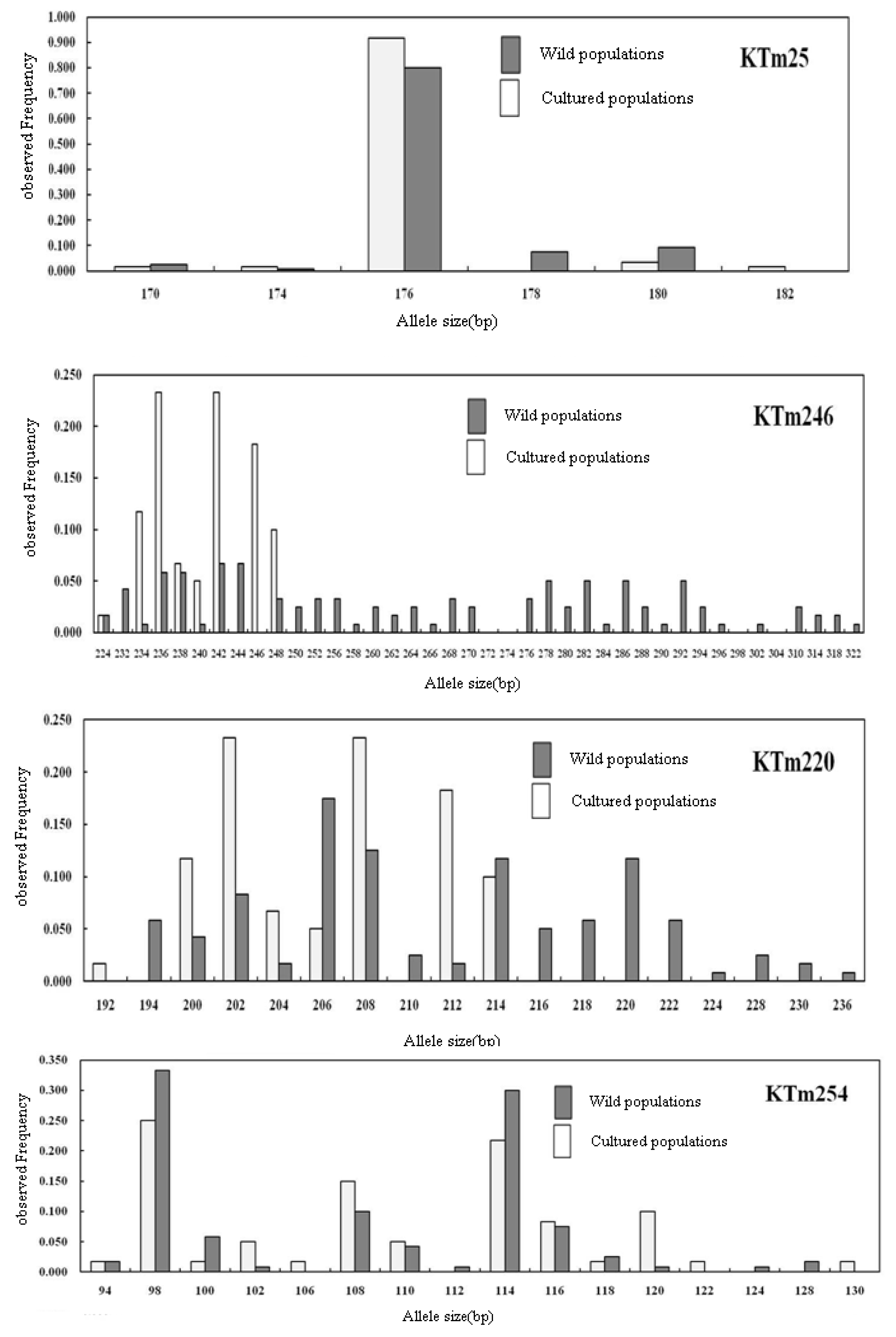

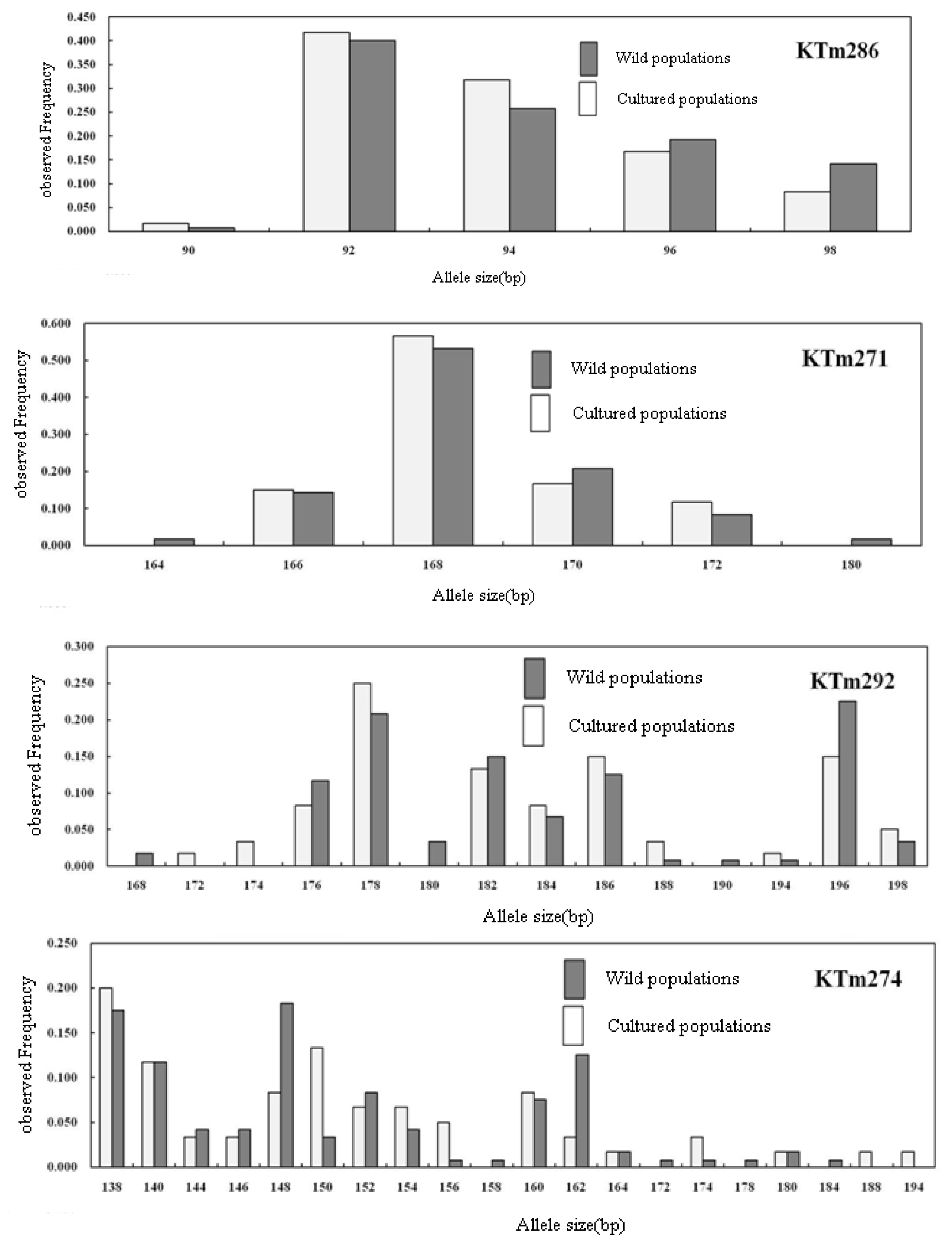
| Locus | Repeat Motif | Primer Sequence (5′→3′) | Ta (°C) | Genbank Accession No. |
|---|---|---|---|---|
| KTm138 | (CA)13-(CA)8 | F: CTAATTTCCCAAGGTAAGGTT 6-fam R: TCATTAAAAAGCCAATCAGAA | 60 | FJ210983 |
| KTm141 | (GT)12 | F: CGTGATGTCGCTGTAAGG hex R: TCTGGCAGTTTTTCTTTTTAT | 57 | FJ210984 |
| KTm151 | (GT)22 | F: CACAGACAGAAGGTCAGAGAG 6-fam R: ATTCATGCAACTTACACGACT | 55 | FJ210987 |
| KTm25 | (GT)6-(GT)5 | F: TGATTACCTCAAAACTTGTGT hex R: CCTTCAGCTGTAAATCTCATA | 57 | FJ210993 |
| KTm220 | (GT)7CT(GT)7AT(GT)10 | F: AGTTTTTAGATTTGCGGTTGT 6-fam R: AATTCCCCCTACAGTCTTCTC | 57 | FJ210997 |
| KTm222 | (GT)13 | F: TGGGTTTTGTTGGGTAA hex R: CCCCCTCTATTTAGATTATGT | 57 | FJ210999 |
| KTm223 | (CA)11CT(CA)9 | F: CTGCTGCACGTGTCTCA ned R: TGTGATAGCCAAAGTCTGATG | 57 | FJ211000 |
| KTm234 | (CA)14 | F: TTGCCTTTACAGTGACAAACA 6-fam R: GGATGGAGGGGAAGAGA | 63 | FJ211001 |
| KTm246 | (CA)12 | F: AAACGGCTCATGTTAATCTGT ned R: AATTCTGCAGCCGTTTAGAC | 63 | FJ211003 |
| KTm252 | (GT)24 | F: ACACGTTTGCAGATTTGTAAT hex R: GCCACTCTCTAGGGTAGTAGG | 57 | FJ211006 |
| KTm254 | (GT)13 | F: AGCCCTAATATAAACACACTG hex R: TGCGCAGGATACATTG | 52 | FJ211007 |
| KTm255 | (GT)11 | F: AGCGAAGAGAACATTCCTC ned R: CTGCCAAGATCCTAACTTTGT | 57 | FJ211008 |
| KTm261 | (GT)17 | F: TTCAGATTTGATTGTGAGGAT 6-fam R: TGAAGGGCAGACTTGTTTAC | 63 | FJ211010 |
| KTm268 | (GT)10 | F: ATTATCACCCCCACAAGTTCT ned R: CGGCTAAATCATGTTTCTGA | 60 | FJ211012 |
| KTm271 | (GT)16 | F: TGTGGTGTTTACTGCAGATAA hex R: ACAGGGTCATGAAAATAATGT | 57 | FJ211013 |
| KTm274 | (GT)20 | F: ATGGAAATAAGCCTCTTGTTC hex R: CTTCTGCAAACCTAAATCAAA | 57 | FJ211016 |
| KTm279 | (GT)5TT(GT)15 | F: AGTTTGACGGCTGACATTTAT hex R: AGCGCTTCAGATCAGATTCT | 57 | FJ211018 |
| KTm283 | (CA)35 | F: GATCTCCATCTCCACCTT 6-fam R: TTGGCTATTGGTAAATTATTC | 57 | FJ211019 |
| KTm286 | (CA)12 | F: GAACTGTGCAACTGTGTTTTC hex R: AAATGTCGCTGTATCTGCTG | 63 | FJ211021 |
| KTm292 | (GT)14 | F: ATCTGCCATTCACTTCCTTTA hex R: CGGAATGTGATCTGCTTTG | 51 | FJ211022 |
| Population (No) | Microsatellite Loci
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KTm 138 | KTm 141 | KTm 151 | KTm 25 | KTm 220 | KTm 222 | KTm 223 | KTm 234 | KTm 246 | KTm 252 | KTm 254 | KTm 255 | KTm 261 | KTm 268 | KTm 271 | KTm 274 | KTm 279 | KTm 283 | KTm 286 | KTm 292 | Mean | ||
| FST | −0.007 | 0.028 | 0.025 | 0.026 | 0.018 | 0.017 | 0.002 | −0.002 | 0.010 | −0.006 | 0.008 | −0.001 | 0.009 | 0.007 | −0.013 | 0.006 | −0.002 | 0.002 | −0.011 | −0.006 | 0.005 | |
| Geoje Wild (60) | NA | 9 | 8 | 13 | 5 | 17 | 13 | 15 | 9 | 35 | 18 | 13 | 16 | 9 | 12 | 6 | 18 | 14 | 20 | 5 | 12 | 12.44 |
| AR | 7.73 | 7.37 | 9.88 | 4.38 | 14.95 | 11.05 | 11.69 | 7.93 | 28.13 | 14.42 | 10.35 | 14.18 | 7.99 | 9.50 | 5.50 | 14.36 | 11.47 | 17.16 | 4.50 | 10.13 | 10.53 | |
| S | 180–208 | 92–108 | 224–270 | 170–180 | 194–236 | 124–154 | 250–286 | 210–230 | 224–322 | 104–144 | 94–128 | 320–368 | 224–240 | 322–370 | 164–180 | 138–184 | 142–192 | 150–218 | 90–98 | 168–198 | ||
| F | 0.442 | 0.375 | 0.342 | 0.800 | 0.175 | 0.258 | 0.408 | 0.342 | 0.067 | 0.175 | 0.333 | 0.258 | 0.433 | 0.333 | 0.533 | 0.183 | 0.300 | 0.175 | 0.400 | 0.225 | 0.32 | |
| U | 3 | 1 | 4 | 1 | 5 | 3 | 5 | 4 | 18 | 4 | 3 | 5 | 2 | 5 | 2 | 4 | 6 | 4 | 0 | 3 | 3.4 | |
| HE | 0.743 | 0.779 | 0.756 | 0.348 | 0.911 | 0.846 | 0.789 | 0.768 | 0.968 | 0.898 | 0.783 | 0.879 | 0.755 | 0.812 | 0.650 | 0.894 | 0.817 | 0.918 | 0.722 | 0.854 | 0.811 | |
| Ho | 0.750 | 0.750 | 0.733 | 0.333 | 0.917 | 0.800 | 0.800 | 0.767 | 0.967 | 0.900 | 0.933 | 0.933 | 0.750 | 0.867 | 0.433 | 0.917 | 0.867 | 0.950 | 0.500 | 0.800 | 0.780 | |
| FIS | −0.010 (0.990) | 0.037 (0.040) | 0.030 (0.044) | 0.043 (0.587) | −0.007 (0.131) | 0.055 (0.724) | −0.015 (0.501) | 0.001 (0.739) | 0.002 (0.525) | −0.002 (0.579) | −0.194 (0.005) | −0.063 (0.302) | 0.006 (0.740) | −0.068 (0.593) | 0.335 (0.000) | −0.025 (0.924) | −0.062 (0.043) | −0.035 (0.650) | 0.310 (0.000) | 0.064 (0.300) | ||
| P | 0.984 | 0.039 | 0.064 | 0.621 | 0.127 | 0.661 | 0.541 | 0.698 | 0.600 | 0.547 | 0.008 | 0.304 | 0.756 | 0.639 | 0.000 | 0.903 | 0.033 | 0.683 | 0.000 | 0.352 | ||
| Geoje Hatchery (30) | NA | 7 | 7 | 11 | 5 | 13 | 10 | 13 | 5 | 22 | 17 | 13 | 12 | 7 | 8 | 4 | 16 | 9 | 18 | 5 | 11 | 10.00 |
| AR | 7.00 | 7.00 | 11.00 | 5.00 | 13.00 | 10.00 | 13.00 | 5.00 | 22.00 | 17.00 | 13.00 | 12.00 | 7.00 | 8.00 | 4.00 | 16.00 | 9.00 | 18.00 | 5.00 | 11.00 | 10.00 | |
| S | 182–194 | 92–106 | 224–250 | 170–182 | 192–230 | 126–152 | 248–282 | 216–224 | 232–304 | 104–146 | 94–130 | 324–368 | 226–240 | 320–338 | 166–172 | 138–194 | 144–190 | 146–210 | 90–98 | 172–198 | ||
| F | 0.500 | 0.433 | 0.233 | 0.917 | 0.167 | 0.333 | 0.367 | 0.383 | 0.117 | 0.200 | 0.250 | 0.317 | 0.433 | 0.233 | 0.567 | 0.200 | 0.367 | 0.150 | 0.417 | 0.250 | 0.33 | |
| U | 1 | 0 | 2 | 1 | 1 | 0 | 3 | 0 | 5 | 3 | 3 | 1 | 0 | 1 | 0 | 2 | 1 | 2 | 0 | 2 | 1.0 | |
| HE | 0.706 | 0.723 | 0.860 | 0.160 | 0.915 | 0.786 | 0.795 | 0.694 | 0.950 | 0.897 | 0.859 | 0.831 | 0.731 | 0.841 | 0.625 | 0.913 | 0.759 | 0.915 | 0.703 | 0.870 | 0.799 | |
| Ho | 0.767 | 0.900 | 0.700 | 0.100 | 0.933 | 0.967 | 0.967 | 0.600 | 0.967 | 0.967 | 0.967 | 0.800 | 0.733 | 0.833 | 0.467 | 0.867 | 0.700 | 0.933 | 0.567 | 0.800 | 0.744 | |
| FIS | −0.087 (0.635) | −0.250 (0.096) | 0.189 (0.090) | 0.381 (0.002) | −0.020 (0.431) | −0.235 (0.003) | −0.220 (0.480) | 0.138 (0.103) | −0.018 (0.603) | −0.080 (0.123) | −0.128 (0.116) | 0.038 (0.524) | −0.004 (0.902) | 0.009 (0.031) | 0.257 (0.187) | 0.052 (0.342) | 0.079 (0.674) | −0.021 (0.010) | 0.196 (0.158) | 0.082 | ||
| P | 0.640 | 0.091 | 0.051 | 0.004 | 0.447 | 0.004 | 0.464 | 0.114 | 0.685 | 0.181 | 0.989 | 0.135 | 0.539 | 0.907 | 0.035 | 0.199 | 0.390 | 0.663 | 0.014 | 0.132 | ||
| Species (No) | Microsatellite Loci
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KTm 141 | KTm 151 | KTm 220 | KTm 222 | KTm 234 | KTm 252 | KTm 254 | KTm 255 | KTm 261 | KTm 274 | KTm 279 | KTm 292 | ||
| Step hanolepis cirrhifer (20) | NA | 6 | 7 | 8 | 7 | 6 | 6 | 4 | 6 | 6 | 7 | 5 | 7 |
| S | 92–106 | 228–244 | 206–228 | 132–186 | 216–230 | 106–130 | 94–120 | 328–368 | 216–240 | 138–154 | 138–170 | 172–196 | |
| Ta (°C) | 52 | 57 | 57 | 52 | 52 | 57 | 52 | 63 | 63 | 57 | 57 | 52 | |
© 2011 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
An, H.S.; Kim, E.M.; Lee, J.W.; Dong, C.M.; Lee, B.I.; Kim, Y.C. Novel Polymorphic Microsatellite Loci for the Korean Black Scraper (Thamnaconus modestus), and Their Application to the Genetic Characterization of Wild and Farmed Populations. Int. J. Mol. Sci. 2011, 12, 4104-4119. https://doi.org/10.3390/ijms12064104
An HS, Kim EM, Lee JW, Dong CM, Lee BI, Kim YC. Novel Polymorphic Microsatellite Loci for the Korean Black Scraper (Thamnaconus modestus), and Their Application to the Genetic Characterization of Wild and Farmed Populations. International Journal of Molecular Sciences. 2011; 12(6):4104-4119. https://doi.org/10.3390/ijms12064104
Chicago/Turabian StyleAn, Hye Suck, Eun Mi Kim, Jang Wook Lee, Chun Mae Dong, Bai Ik Lee, and Yi Cheong Kim. 2011. "Novel Polymorphic Microsatellite Loci for the Korean Black Scraper (Thamnaconus modestus), and Their Application to the Genetic Characterization of Wild and Farmed Populations" International Journal of Molecular Sciences 12, no. 6: 4104-4119. https://doi.org/10.3390/ijms12064104




