A Computational Simulation Study of Benzamidine Derivatives Binding to Arginine-Specific Gingipain (HRgpA) from Periodontopathogen Porphyromonas gingivalis
Abstract
:1. Introduction
2. Results and Discussion
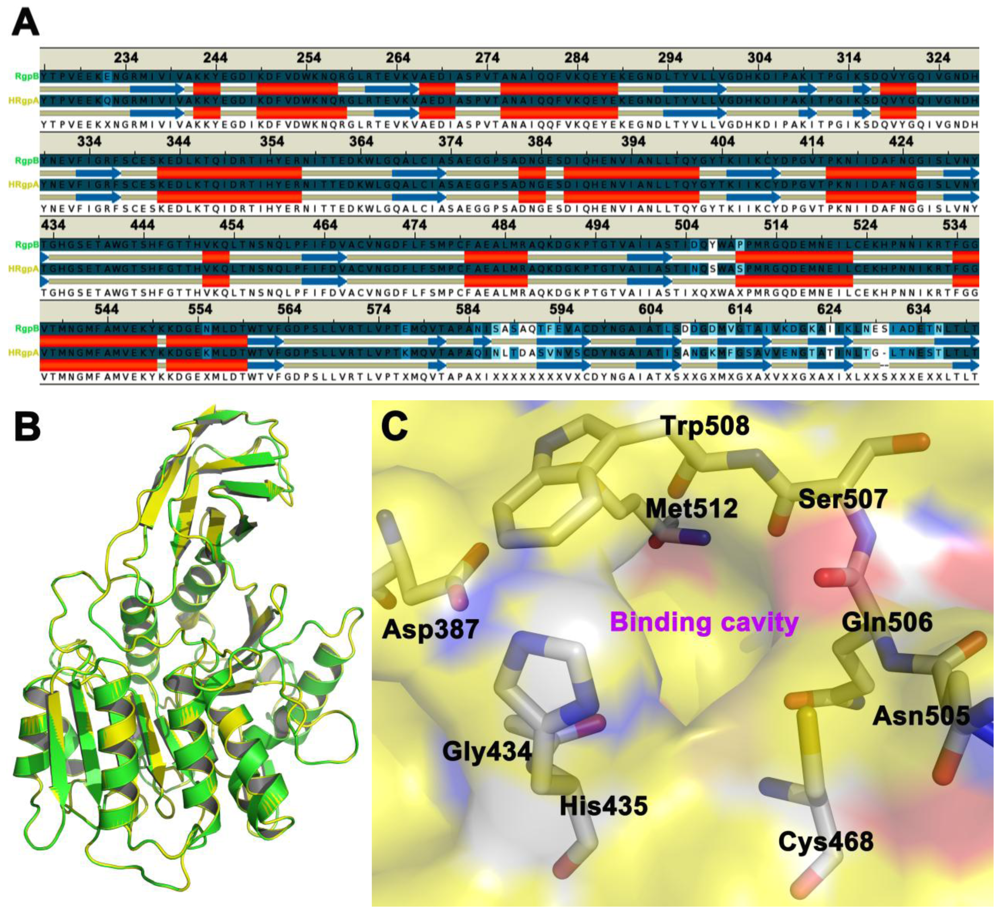
2.1. Homology Modeling
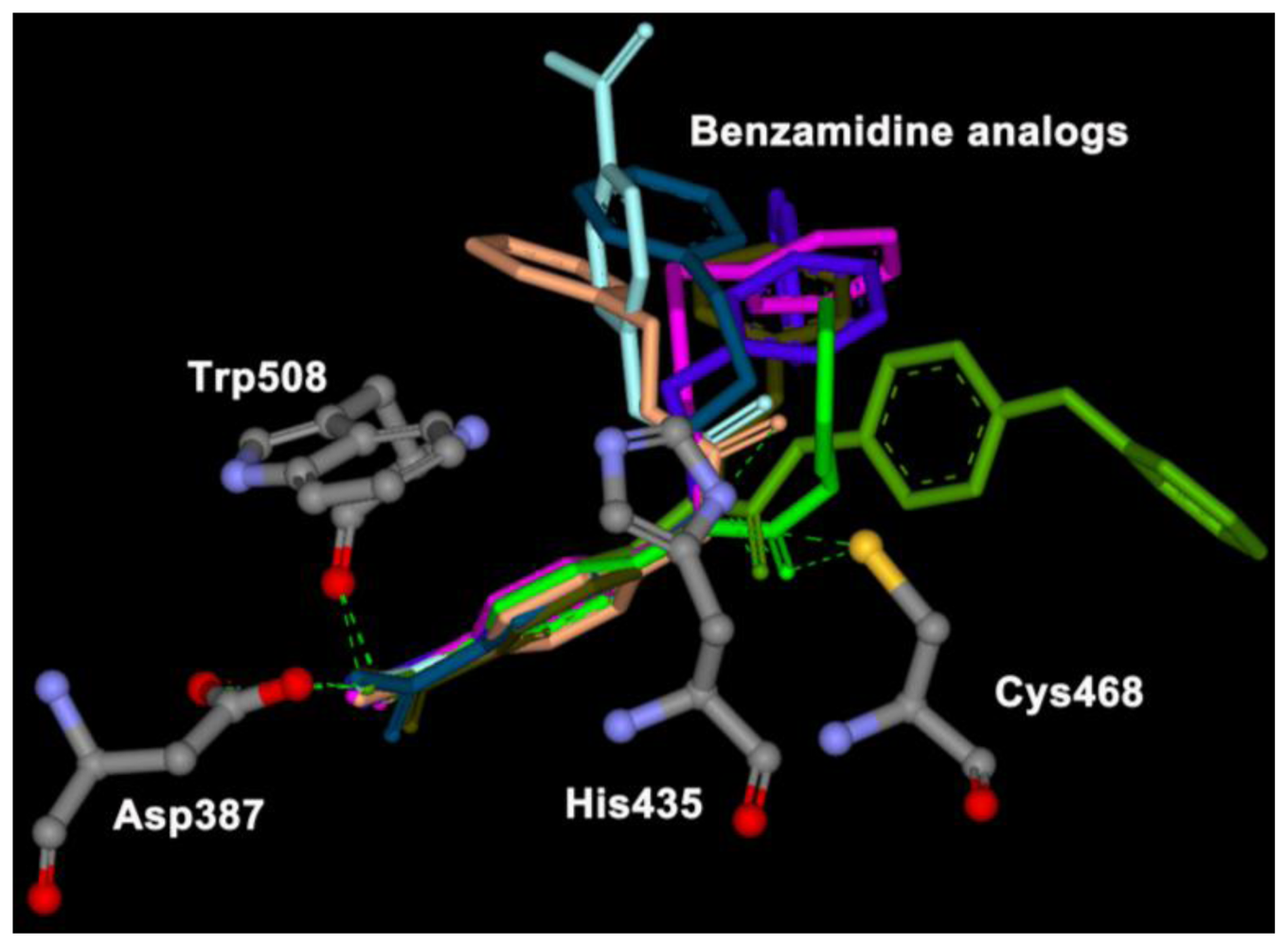
2.2. Automated Docking
2.3. Molecular Dynamics
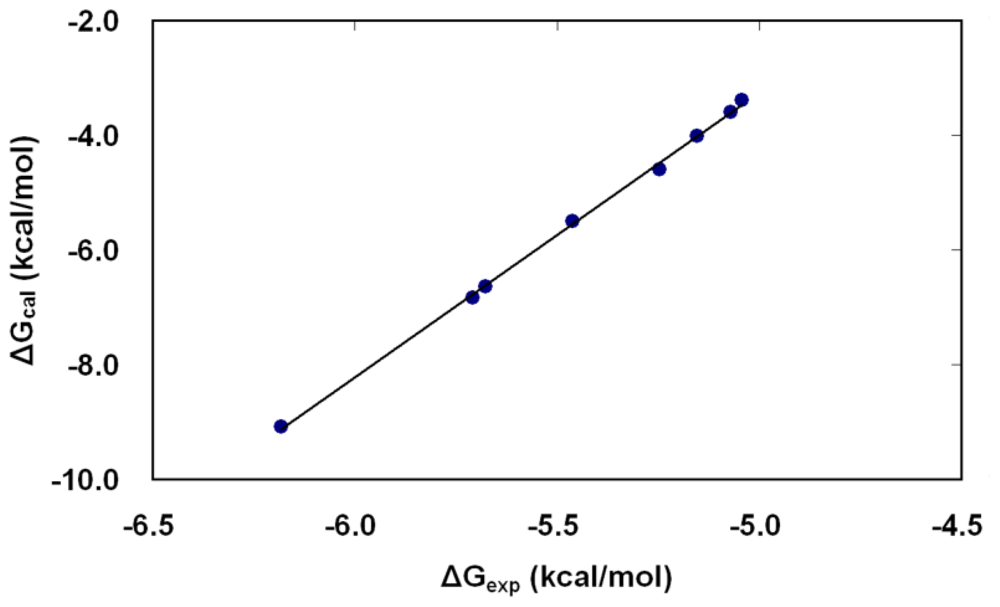
2.4. Binding Free Energy
3. Computational Methods
3.1. Homology Modeling and Docking
3.2. Molecular Dynamics Simulation
3.3. Free Energy Calculation
4. Conclusions
Acknowledgements
References
- Imamura, T. The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol 2003, 74, 111–118. [Google Scholar]
- Sheets, SM; Robles-Price, AG; McKenzie, RM; Casiano, CA; Fletcher, HM. Gingipaindependent interactions with the host are important for survival of Porphyromonas gingivalis. Front Biosci 2008, 13, 3215–3238. [Google Scholar]
- Kadowaki, T; Takii, R; Yamatake, K; Kawakubo, T; Tsukuba, T; Yamamoto, K. A role for gingipains in cellular responses and bacterial survival in Porphyromonas gingivalis-infected cells. Front Biosci 2007, 12, 4800–4809. [Google Scholar]
- Potempa, J; Banbula, A; Travis, J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol. 2000 2000, 32, 153–192. [Google Scholar]
- Travis, J; Banbula, A; Potempa, J. The role of bacterial and host proteinases in periodontal disease. Adv. Exp. Med. Biol 2000, 477, 455–465. [Google Scholar]
- Ellen, RP; Lépine, G; Nghiem, PM. In vitro models that support adhesion specificity in biofilms of oral bacteria. Adv. Dent. Res 1997, 11, 33–42. [Google Scholar]
- Fitzpatrick, RE; Wijeyewickrema, LC; Pike, RN. The gingipains: Scissors and glue of the periodontal pathogen, Porphyromonas gingivalis. Future Microbiol 2009, 4, 471–487. [Google Scholar]
- Yamamoto, K; Kadowaki, T; Okamoto, K; Yoneda, M; Nakayama, K; Misumi, Y; Ikehara, Y. Structure and function of a novel arginine-specific cysteine proteinase (argingipain) as a major periodontal pathogenic factor from porphyromonas gingivalis. Adv. Exp. Med. Biol 1996, 389, 33–42. [Google Scholar]
- Nakayama, K. Molecular genetics of Porphyromonas gingivalis: Gingipains and other virulence factors. Curr. Protein Pept. Sci 2003, 4, 389–395. [Google Scholar]
- Potempa, J; Sroka, A; Imamura, T; Travis, J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: Structure, function and assembly of multidomain protein complexes. Curr. Protein Pept. Sci 2003, 4, 397–407. [Google Scholar]
- Curtis, MA; Aduse-Opoku, J; Rangarajan, M. Cysteine proteases of Porphyromonas gingivalis. Crit. Rev. Oral. Biol. Med 2001, 12, 192–216. [Google Scholar]
- Genco, CA; Potempa, J; Mikolajczyk-Pawlinska, J; Travis, J. Role of gingipains R in the pathogenesis of Porphyromonas gingivalis–mediated periodontal disease. Clin. Infect. Dis 1999, 28, 456–465. [Google Scholar]
- Kadowaki, T; Nakayama, K; Okamoto, K; Abe, N; Baba, A; Shi, Y; Ratnayake, DB; Yamamoto, K. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J. Biochem 2000, 128, 153–159. [Google Scholar]
- Krauser, JA; Potempa, J; Travis, J; Powers, JC. Inhibition of arginine gingipains (RgpB and HRgpA) with benzamidine inhibitors: Zinc increases inhibitory potency. Biol. Chem 2002, 383, 1193–1198. [Google Scholar]
- Eichinger, A; Beisel, HG; Jacob, U; Huber, R; Medrano, FJ; Banbula, A; Potempa, J; Travis, J; Bode, W. Crystal structure of gingipain R: An Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J 1999, 18, 5453–5462. [Google Scholar]
- Thomas, AH. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comp. Chem 1996, 17, 490–519. [Google Scholar]
- Koska, J; Spassov, VZ; Maynard, AJ; Yan, L; Austin, N; Flook, PK; Venkatachalam, CM. Fully automated molecular mechanics based induced fit protein-ligand docking method. J. Chem. Inf. Model 2008, 48, 1965–1973. [Google Scholar]
- Venkatachalam, CM; Jiang, X; Oldfield, T; Waldman, M. LigandFit: A novel method for the shape-directed rapid docking of ligands to protein active sites. J. Mol. Graphics Modell 2003, 21, 289–307. [Google Scholar]
- Case, DA; Darden, TA; Cheatham, TE, III; Simmerling, CL; Wang, J; Duke, RE; Luo, R; Crowley, M; Walker, RC; Zhang, W; et al. AMBER 10, Users’ Manual; University of California: San Francisco, CA, USA, 2008. [Google Scholar]
- Case, DA; Cheatham, TE, III; Darden, T; Gohlke, H; Luo, R; Merz, KM, Jr; Onufriev, A; Simmerling, C; Wang, B; Woods, R. The Amber biomolecular simulation programs. J. Computat. Chem 2005, 26, 1668–1688. [Google Scholar]
- Ponder, JW; Case, DA. Force fields for protein simulations. Adv. Prot. Chem 2003, 66, 27–85. [Google Scholar]
- Cheatham, TE, III; Young, MA. Molecular dynamics simulation of nucleic acids: Successes, limitations and promise. Biopolymers 2001, 56, 232–256. [Google Scholar]
- Jorgensen, WL; Chandrasekhar, J; Madura, JD; Impey, RW; Klein, ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 1983, 79, 926–935. [Google Scholar]
- King, G; Warshel, A. A surface constrained all-atom solvent model for effective simulations of polar solutions. J. Chem. Phys 1989, 91, 3647–3661. [Google Scholar]
- Ryckaert, JP; Ciccotti, G; Berendsen, HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys 1977, 23, 327–341. [Google Scholar]
- Lee, FS; Warshel, A. A local reaction field method for fast evaluation of long-range electrostatic interactions in molecular simulations. J. Chem. Phys 1992, 97, 3100–3107. [Google Scholar]
- Chen, SWW; Honig, B. Monovalent and divalent salt effects on electrostatic free energies defined by the nonlinear poisson-boltzmann equation: Application to DNA recognition. J. Phys. Chem. B 1997, 101, 9113–9118. [Google Scholar]
- Honig, B; Nicholls, A. Classical electrostatics in biology and chemistry. Science 1995, 268, 1144–1149. [Google Scholar]
- Sharp, KA; Honig, B. Electrostatic interactions in macromolecules: Theory and application. Annu. Rev. Biophys. Biophys. Chem 1990, 19, 301–332. [Google Scholar]
- Madura, JD; Davis, ME; Gilson, MK; Wade, RC; Luty, BA; McCammon, JA. Lipkowitz, KB, Boyd, DB, Eds.; Biological applications of electrostatic calculations and brownian dynamics simulations. In Reviews in Computational Chemistry; VCH: New York, NY, USA, 1994; Volume 5, pp. 229–267. [Google Scholar]
- Davis, ME; McCammon, JA. Electrostatics in biomolecular structure and dynamics. Chem. Rev 1990, 90, 509–521. [Google Scholar]
- Gilson, MK; Honig, B. Calculation of the total electrostatic energy of a macromolecular system: Solvation energies, binding energies, and conformational analysis. Proteins 1988, 4, 7–18. [Google Scholar]
- Cramer, CJ; Truhlar, DG. Lipkowitz, KB, Boyd, DB, Eds.; Continuum solvation models: Classical and quantum mechanical implementation. In Reviews in Computational Chemistry; VCH: New York, NY, USA, 1995; Volume 6, pp. 1–72. [Google Scholar]
- Sitkoff, D; Sharp, KA; Honig, B. Correlating solvation free energies and surface tensions of hydrocarbons. Biophys. Chem 1994, 51, 397–409. [Google Scholar]
- Hermann, RB. Use of solvent cavity area and number of packed solvent molecules around a solute in regard to hydrocarbon solubilities and hydrophobic interactions. Proc. Natl. Acad. Sci. USA 1977, 74, 4144–4145. [Google Scholar]
- Honig, B; Sharp, K; Yang, AS. Macroscopic models of aqueous solutions: Biological and chemical applications. J. Phys. Chem 1993, 97, 1101–1109. [Google Scholar]
- Tunon, I; Silla, E; Pascualahuir, JL. Molecular surface area and hydrophobic effect. Protein Eng 1992, 5, 715–716. [Google Scholar]
- Ooi, T; Oobatake, M; Nemethy, G; Scheraga, HA. Accessible surface areas as a measure of the thermodynamic parameters of hydration of peptides. Proc. Natl. Acad. Sci. USA 1987, 84, 3086–3090. [Google Scholar]
- Rashin, AA; Bukatin, MA. A view of thermodynamics of hydration emerging from continuum studies. Biophys. Chem 1994, 51, 167–192. [Google Scholar]


| Name | Compounds | Ki (μM) | ΔGexp |
|---|---|---|---|
| Bz1 | 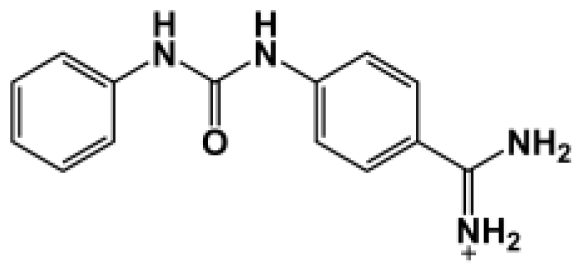 | 68.3 | −5.676 |
| Bz2 |  | 98.1 | −5.461 |
| Bz3 |  | 64.7 | −5.708 |
| Bz4 |  | 29.0 | −6.183 |
| Bz5 | 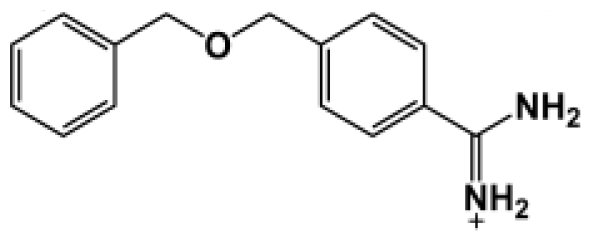 | 141.0 | −5.247 |
| Bz6 | 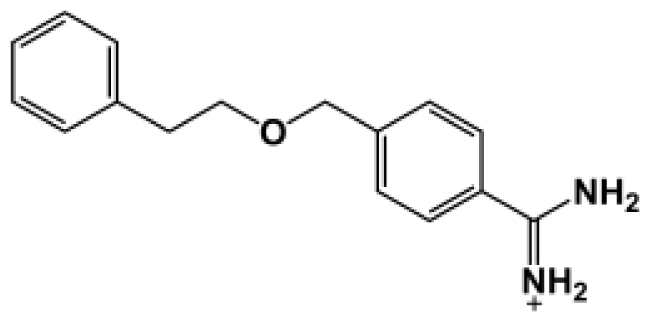 | 165.0 | −5.154 |
| Bz7 | 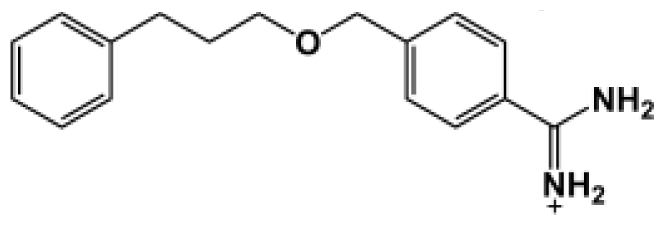 | 190.0 | −5.070 |
| Bz8 | 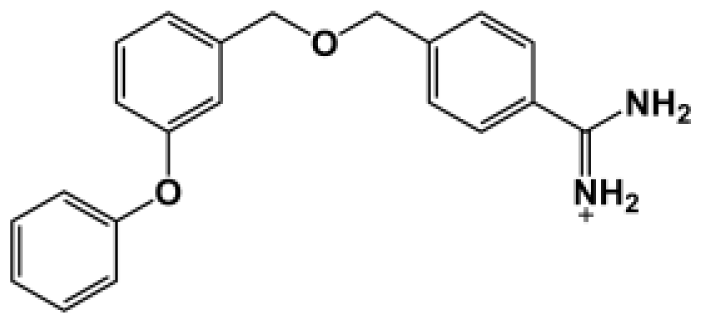 | 199.0 | −5.043 |
| Name | ΔGelec | ΔGvdW | ΔGnonp/sol | ΔGelec/sol | −TΔS | ΔGbind |
|---|---|---|---|---|---|---|
| Bz1 | −25.89 ± 0.09 | −13.88 ± 0.12 | −3.42 ± 0.01 | 24.14 ± 0.01 | 12.36 ± 0.28 | −6.69 ± 0.10 |
| Bz2 | −25.11 ± 0.01 | −13.81 ± 0.12 | −3.45 ± 0.01 | 24.24 ± 0.03 | 12.61 ± 0.54 | −5.52 ± 0.14 |
| Bz3 | −25.21 ± 0.11 | −14.33 ± 0.12 | −3.31 ± 0.01 | 23.23 ± 0.01 | 12.79 ± 0.31 | −6.83 ± 0.11 |
| Bz4 | −26.21 ± 0.10 | −15.02 ± 0.13 | −3.28 ± 0.01 | 23.05 ± 0.04 | 12.38 ± 0.63 | −9.08 ± 0.18 |
| Bz5 | −24.26 ± 0.11 | −13.52 ± 0.07 | −3.34 ± 0.01 | 23.51 ± 0.02 | 13.63 ± 0.55 | −4.68 ± 0.15 |
| Bz6 | −26.03 ± 0.10 | −13.37 ± 0.16 | −3.18 ± 0.01 | 24.31 ± 0.02 | 14.25 ± 0.39 | −4.02 ± 0.14 |
| Bz7 | −25.55 ± 0.08 | −13.78 ± 0.11 | −3.45 ± 0.01 | 24.61 ± 0.05 | 14.57 ± 0.71 | −3.60 ± 0.12 |
| Bz8 | −25.16 ± 0.09 | −13.03 ± 0.13 | −3.04 ± 0.01 | 24.15 ± 0.03 | 13.69 ± 0.59 | −3.39 ± 0.17 |
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kim, D.; Lee, D.-S. A Computational Simulation Study of Benzamidine Derivatives Binding to Arginine-Specific Gingipain (HRgpA) from Periodontopathogen Porphyromonas gingivalis. Int. J. Mol. Sci. 2010, 11, 3252-3265. https://doi.org/10.3390/ijms11093252
Kim D, Lee D-S. A Computational Simulation Study of Benzamidine Derivatives Binding to Arginine-Specific Gingipain (HRgpA) from Periodontopathogen Porphyromonas gingivalis. International Journal of Molecular Sciences. 2010; 11(9):3252-3265. https://doi.org/10.3390/ijms11093252
Chicago/Turabian StyleKim, Dooil, and Dae-Sil Lee. 2010. "A Computational Simulation Study of Benzamidine Derivatives Binding to Arginine-Specific Gingipain (HRgpA) from Periodontopathogen Porphyromonas gingivalis" International Journal of Molecular Sciences 11, no. 9: 3252-3265. https://doi.org/10.3390/ijms11093252




