Production and Characterization of Fengycin by Indigenous Bacillus subtilis F29-3 Originating from a Potato Farm
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of Fengycin
2.1.1. IR Spectrometric Analyses
2.1.2. UV Spectrometric Analyses
2.1.3. NMR Spectrometric Analyses
2.1.4. MALDI-TOF/MASS Analyses
2.2. Optimization of Medium Constituents for Fengycin Production by RSM
2.2.1. Fractional Factorial Design
2.2.2. Steepest Ascent Method
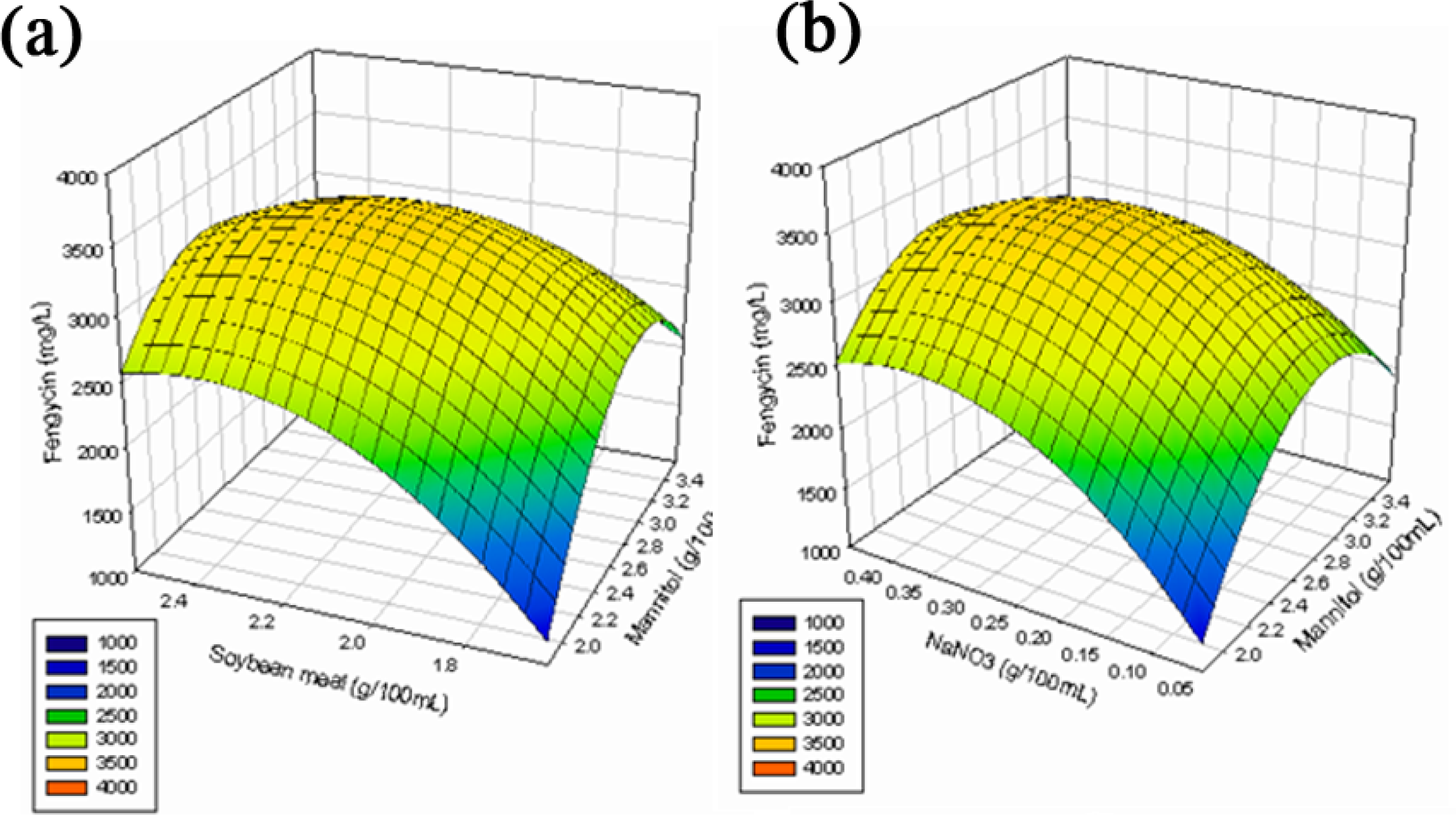
2.2.3. Response Surface Methodology (RSM)
3. Experimental Section
3.1. Microorganism
3.2. Growth Medium and Culture Conditions
3.3. Purification of Fengycin
3.4. Characterization of Fengycin
3.4.1. Fast Atom Bombardment-Mass Spectrometry (FAB-MS)
3.4.2. IR Spectrometric Analyses
3.4.3. UV Spectrometric Analyses
3.4.4. NMR Analysis
3.4.5. MALDI-TOF/MASS
3.5. Quantification of Fengycin
3.6. Optimization of Medium Composition for Fengycin Production
3.6.1. Fractional Factorial Design
3.6.2. Steepest Ascent Method
3.6.3. Response Surface Methodology (RSM)
4. Conclusions
Supplementary Material
ijms-11-04526-s001.pdfAcknowledgments
References
- Cameotra, SS; Makkar, RS; Kaur, J; Mehta, SK. Synthesis of biosurfactants and their advantages to microorganisms and mankind. Adv. Exp. Med. Biol 2010, 672, 261–280. [Google Scholar]
- Baker, SC; Chen, CY. Enrichment and purification of lipopeptide biosurfactants. Adv. Exp. Med. Biol 2010, 672, 281–288. [Google Scholar]
- Lourith, N; Kanlayavattanakul, M. Natural surfactants used in cosmetics: glycolipids. Int. J. Cosmet. Sci 2009, 31, 255–261. [Google Scholar]
- Vater, J; Kablitz, B; Wilde, C; Franke, P; Mehta, N; Cameotra, SS. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry of Lipopeptide Biosurfactants in Whole Cells and Culture Filtrates of Bacillus subtilis C-1 Isolated from Petroleum Sludge. Appl. Environ. Microbiol 2002, 68, 6210–6219. [Google Scholar]
- Wei, YH; Chu, IM. Enhancement of surfactin production in iron-enriched media by Bacillus subtilis ATCC 21332. Enzyme Microbial. Technol 1998, 22, 724–728. [Google Scholar]
- Chen, SY; Lu, WB; Wei, YH; Chen, WM; Chang, JS. Improved production of biosurfactant with newly isolated Pseudomonas aeruginosa S2. Biotechnol. Prog 2007, 23, 661–666. [Google Scholar]
- Wei, YH; Lai, HC; Chen, SY; Yeh, MS; Chang, JS. Biosurfactant production by Serratia marcescens SS-1 and its isogenic strain SMdeltaR defective in SpnR, a quorum-sensing LuxR family protein. Biotechnol. Lett 2004, 26, 799–802. [Google Scholar]
- Perfumo, A; Rancich, I; Banat, IM. Possibilities and challenges for biosurfactants use in petroleum industry. Adv. Exp. Med. Biol 2010, 672, 135–145. [Google Scholar]
- Sen, R. Surfactin: biosynthesis, genetics and potential applications. Adv. Exp. Med. Biol 2010, 672, 316–323. [Google Scholar]
- Steller, S; Vollenbroich, D; Leenders, F; Stein, T; Conrad, B; Hofemeister, J; Jacques, P; Thonart, P; Vater, J. Structural and functional organization of the fengycin synthetase multienzyme system from Bacillus Subtilis b213 and A1/3. Chem. Biol 1999, 6, 31–41. [Google Scholar]
- Wei, YH; Wang, LF; Chang, JS; Kung, SS. Identification of induced acidification in iron-enriched cultures of Bacillus subtilis during biosurfactant fermentation. J. Biosci. Bioeng 2003, 96, 174–178. [Google Scholar]
- Steller, S; Vater, J. Purification of the fengycin synthetase multienzyme system from Bacillus subtilis b213. J. Chromato. Biomed. Sci 2000, 737, 267–275. [Google Scholar]
- Loeffler, W; Tschen, JSM; Vanittanakom, N; Kugler, M; Knorpp, E; Hsieh, TF; Wu, TG. Antifungal Effects of Bacilysin and Fengymycin from Bacillus subtilis F-29-3 A comparsion with Activities of Other Bacillus Antibiotics. J. Phytopath 1986, 115, 204–213. [Google Scholar]
- Vanittanakom, N; Loeffler, W; Koch, U; Jung, G. Fengycin—a novel antifungal lipopeptide antibiotic produced by Bacillus Subtilis F-29-3. J. Antibiot 1986, 39, 888–901. [Google Scholar]
- Lin, SC; Minton, MA; Sharma, MM; Georgiou, G. Structural and immunological characterization of a biosurfactant produced by Bacillus licheniformis JF-2. Appl. Environ. Microbiol 1994, 60, 31–38. [Google Scholar]
- Ke, WJ; Chang, BY; Lin, TP; Liu, ST. Activation of the promoter of the fengycin synthetase operon by the UP element. J. Bacteriol 2009, 191, 4615–4623. [Google Scholar]
- Lin, TP; Chen, CL; Fu, HC; Wu, CY; Lin, GH; Huang, SH; Chang, LK; Liu, ST. Functional analysis of fengycin synthetase FenD. Biochim. Biophys. Acta 2005, 1730, 159–164. [Google Scholar]
- Wei, YH; Wang, LF; Chang, JS. Optimizing iron supplement strategies for enhanced surfactin production with Bacillus subtilis. Biotechnol. Prog 2004, 20, 979–983. [Google Scholar]
- Sun, Y; Han, B; Liu, W; Zhang, J; Gao, X. Substrate induction and statistical optimization for the production of chitosanase from Microbacterium sp. OU01. Bioresour. Technol 2006, 98, 1548–1553. [Google Scholar]
- Kumar, P; Satyanarayana, T. Optimization of culture variables for improving glucoamylase production by alginate-entrapped Thermomucor indicae-seudaticae using statistical methods. Bioresour. Technol 2007, 98, 1252–1259. [Google Scholar]
- Donato, P; Stancanelli, R; Calabro, ML; Tommasini, S; Cutroneo, P; Guardo, M; Pagano, B; Chimirri, A; Ficarra, P; Ficarra, R. Optimization of a LC method for the enantioseparation of a non-competitive glutamate receptor antagonist, by experimental design methodology. J. Pharm. Biomed. Anal 2006, 42, 543–548. [Google Scholar]
- Sen, R. Response Surface Optimization of the Critical Media Components for the Production of Surfactin. J. Chem. Technol. Biotechnol 1997, 68, 263–270. [Google Scholar]
- Pal, MP; Vaidya, BK; Desai, KM; Joshi, RM; Nene, SN; Kulkarni, BD. Media optimization for biosurfactant production by Rhodococcus erythropolis MTCC 2794: artificial intelligence versus a statistical approach. J. Ind. Microbiol. Biotechnol 2009, 36, 747–756. [Google Scholar]

| Retention Time (min) | Main MALDI-TOF Peak(s) (m/z) | Assignment |
|---|---|---|
| 5, 6 | 1523.865, 1509.855 | B-C16 and C17 fengycin [M + Na]+ |
| 6, 7 | 1509.855, 1477.828, 1491.825 | B-C16 fengycin [M + Na]+ |
| A-C17 fengycin [M + H]+ | ||
| B-C16 fengycin [M + H]+ | ||
| 7, 8 | 1491.825, 1505.851 | B-C16 and C17 fengycin [M + H]+ |
| 8, 9 | 1505.898, 1527.901 | B-C17 fengycin [M + H]+ |
| B-C17 fengycin [M + Na]+ | ||
| 9, 10 | 1475.844 | A-C17 fengycin [M + H]+ |
| 10, 11 | 1475.852, 1497.859 | A-C17 fengycin [M + H]+ |
| A-C17 fengycin [M + Na]+ | ||
| 11, 12 | 1475.817, 1497.816 | A-C17 fengycin [M + H]+ |
| A-C17 fengycin [M + Na]+ | ||
| 12, 13 | 1475.793, 1505.808 | A-C17 fengycin [M + H]+ |
| B-C17 fengycin [M + H]+ | ||
| 13, 14 | 1511.853 | B-C16 fengycin [M + Na]+ |
| 14, 15 | 1489.836 | B-C16 fengycin [M + H]+ |
| 15, 16 | 1489.912 | B-C16 fengycin [M + H]+ |
| Run No. | Variables | |||||||
|---|---|---|---|---|---|---|---|---|
| Mannitol (g/100 mL) | Soybean Meal (g/100 mL) | NaNO3 (g/100 mL) | FeCl2·4H2O (g/100 mL) | MgSO4·7H2O (g/100 mL) | MnSO4·4H2O (g/100 mL) | Na2MoO4 (g/100 mL) | Fengycin Production (mg/L) | |
| 1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | 337 ± 31 |
| 2 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1161± 104 |
| 3 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 708 ± 63 |
| 4 | −1 | −1 | 1 | 1 | −1 | −1 | 1 | 542 ± 72 |
| 5 | −1 | 1 | −1 | −1 | 1 | −1 | 1 | 447 ± 51 |
| 6 | −1 | 1 | −1 | 1 | −1 | 1 | −1 | 1688 ± 137 |
| 7 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1066 ± 101 |
| 8 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 644 ± 75 |
| 9 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 1712 ± 148 |
| 10 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 1598 ± 193 |
| 11 | 1 | −1 | 1 | −1 | 1 | −1 | 1 | 1054 ± 119 |
| 12 | 1 | −1 | 1 | 1 | −1 | 1 | −1 | 1527 ± 124 |
| 13 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 2311 ±254 |
| 14 | 1 | 1 | −1 | 1 | −1 | −1 | 1 | 2527 ± 285 |
| 15 | 1 | 1 | 1 | −1 | −1 | −1 | −1 | 1556 ± 199 |
| 16 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1853 ± 162 |
| Source | DF | Sum of Squares | F-Ratio | Prob > F |
|---|---|---|---|---|
| Model | 7 | 5925107.9 | 11.7 | 0.0012 |
| Mannitol | 1 | 3557939.1 | 49.1 | 0.0001 |
| Soybean meal | 1 | 745200.6 | 10.3 | 0.0125 |
| NaNO3 | 1 | 500910.1 | 6.9 | 0.0302 |
| FeCl2·4H2O | 1 | 344862.6 | 4.8 | 0.0606 |
| MgSO4·7H2O | 1 | 86877.6 | 1.2 | 0.3052 |
| MnSO4·4H2O | 1 | 689315.1 | 9.5 | 0.0150 |
| Na2MoO4 | 1 | 3.1 | 0.0 | 0.9950 |
| Experiment No. | Mannitol (g/100 mL) | Soybean Meal (g/100 mL) | NaNO3 (g/100 mL) | MnSO4·4H2O (g/100 mL) |
|---|---|---|---|---|
| 4 | 3.2 | 2.3 | 0.2 | 0.01 |
| 3 | 2.7 | 2.1 | 0.3 | 0.02 |
| 2 | 2.3 | 1.9 | 0.4 | 0.03 |
| 1 | 1.8 | 1.6 | 0.5 | 0.04 |
| 0 | 1.4 | 1.4 | 0.6 | 0.05 |
| −1 | 1.0 | 1.2 | 0.7 | 0.06 |
| −2 | 0.5 | 0.9 | 0.8 | 0.07 |
| Run No. | Mannitol (g/100 mL) | Soybean Meal (g/100 mL) | NaNO3 (g/100 mL) | MnSO4·4H2O (g/100 mL) | Fengycin Production (mg/L) | |
|---|---|---|---|---|---|---|
| Experimental | Predicted | |||||
| 1 | −1 | −1 | −1 | −1 | 3033 ± 313 | 2956 ± 315 |
| 2 | −1 | −1 | −1 | 1 | 2394 ± 259 | 2517 ± 281 |
| 3 | −1 | −1 | 1 | −1 | 2461 ± 216 | 2218 ± 215 |
| 4 | −1 | −1 | 1 | 1 | 1981 ± 238 | 1941 ± 221 |
| 5 | −1 | 1 | −1 | −1 | 3351 ± 315 | 3327 ± 381 |
| 6 | −1 | 1 | −1 | 1 | 2699 ± 289 | 2807 ± 252 |
| 7 | −1 | 1 | 1 | −1 | 2682 ± 278 | 2909 ± 264 |
| 8 | −1 | 1 | 1 | 1 | 2623 ± 242 | 2550 ± 281 |
| 9 | 1 | −1 | −1 | −1 | 2867 ± 236 | 2938 ± 312 |
| 10 | 1 | −1 | −1 | 1 | 2968 ± 256 | 2695 ± 261 |
| 11 | 1 | −1 | 1 | −1 | 2613 ± 281 | 2516 ± 274 |
| 12 | 1 | −1 | 1 | 1 | 2414 ± 261 | 2435 ± 253 |
| 13 | 1 | 1 | −1 | −1 | 2858 ± 275 | 2909 ± 287 |
| 14 | 1 | 1 | −1 | 1 | 2343 ± 284 | 2584 ± 261 |
| 15 | 1 | 1 | 1 | −1 | 2933 ± 213 | 2807 ± 271 |
| 16 | 1 | 1 | 1 | 1 | 2554 ± 215 | 2643 ± 284 |
| 17 | 0 | 0 | 0 | 0 | 3263 ± 326 | 3371 ± 391 |
| 18 | 0 | 0 | 0 | 0 | 3418 ± 321 | 3371 ± 337 |
| 19 | 0 | 0 | 0 | 0 | 3297 ± 323 | 3371 ± 312 |
| 20 | 0 | 0 | 0 | 0 | 3449 ± 324 | 3371 ± 353 |
| 21 | −2 | 0 | 0 | 0 | 2413 ± 211 | 2416 ± 252 |
| 22 | 2 | 0 | 0 | 0 | 2506 ± 230 | 2492 ± 240 |
| 23 | 0 | −2 | 0 | 0 | 2302 ± 220 | 2535 ± 311 |
| 24 | 0 | 2 | 0 | 0 | 2242 ± 254 | 3115 ± 335 |
| 25 | 0 | 0 | −2 | 0 | 2375 ± 257 | 3154 ± 291 |
| 26 | 0 | 0 | 2 | 0 | 2352 ± 215 | 2476 ± 245 |
| 27 | 0 | 0 | 0 | −2 | 2782 ± 248 | 2894 ± 281 |
| 28 | 0 | 0 | 0 | 2 | 2414 ± 261 | 2291 ± 322 |
| 29 | 0 | 0 | 0 | 0 | 3379 ± 357 | 3371 ± 352 |
| 30 | 0 | 0 | 0 | 0 | 3425 ± 322 | 3371 ± 336 |
| Source | Coefficient | Standard Error | t-Value | Prob > t |
|---|---|---|---|---|
| Intercept | 3371.8 | 74.6 | 45.2 | <0.0001 |
| X1 | 18.958 | 37.3 | 0.5 | 0.6184 |
| X2 | 145.1 | 37.3 | 3.9 | 0.0014 |
| X3 | −169.5 | 37.3 | −4.6 | 0.0004 |
| X4 | −150.6 | 37.3 | −4.0 | 0.0011 |
| X1*X1 | −229.3 | 34.9 | −6.6 | <0.0001 |
| X2*X1 | −100.2 | 45.7 | −2.2 | 0.0444 |
| X2*X2 | −136.6 | 34.9 | −3.9 | 0.0014 |
| X3*X1 | 79.1 | 45.7 | 1.7 | 0.1038 |
| X3*X2 | 79.8 | 45.7 | 1.8 | 0.1009 |
| X3*X3 | −139.1 | 34.9 | −4.0 | 0.0012 |
| X4*X1 | 48.8 | 45.7 | 1.1 | 0.3019 |
| X4*X2 | −20.7 | 45.7 | −0.5 | 0.6569 |
| X4*X3 | 40.3 | 45.7 | 0.9 | 0.3912 |
| X4*X4 | −194.7 | 34.9 | −5.6 | <0.0001 |
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wei, Y.-H.; Wang, L.-C.; Chen, W.-C.; Chen, S.-Y. Production and Characterization of Fengycin by Indigenous Bacillus subtilis F29-3 Originating from a Potato Farm. Int. J. Mol. Sci. 2010, 11, 4526-4538. https://doi.org/10.3390/ijms11114526
Wei Y-H, Wang L-C, Chen W-C, Chen S-Y. Production and Characterization of Fengycin by Indigenous Bacillus subtilis F29-3 Originating from a Potato Farm. International Journal of Molecular Sciences. 2010; 11(11):4526-4538. https://doi.org/10.3390/ijms11114526
Chicago/Turabian StyleWei, Yu-Hong, Li-Chuan Wang, Wei-Chuan Chen, and Shan-Yu Chen. 2010. "Production and Characterization of Fengycin by Indigenous Bacillus subtilis F29-3 Originating from a Potato Farm" International Journal of Molecular Sciences 11, no. 11: 4526-4538. https://doi.org/10.3390/ijms11114526




