Fluorescence Lifetime Imaging of Quantum Dot Labeled DNA Microarrays
Abstract
:1. Introduction
2. Experimental Section
2.1. Microarray production and incubation
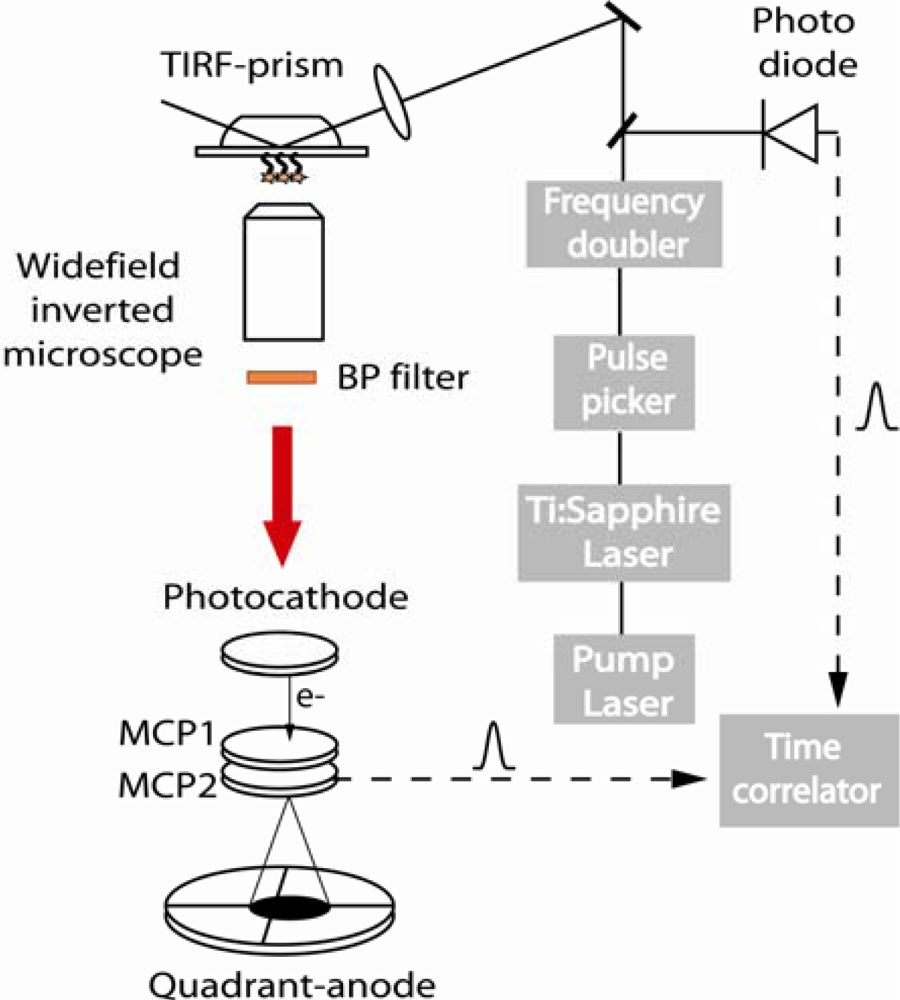
2.2. TIRF-FLIM experimental setup
2.3. Image acquisition and analysis
3. Results and Discussion
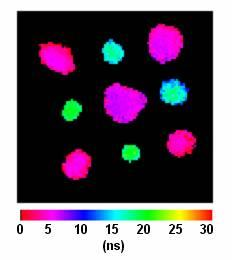
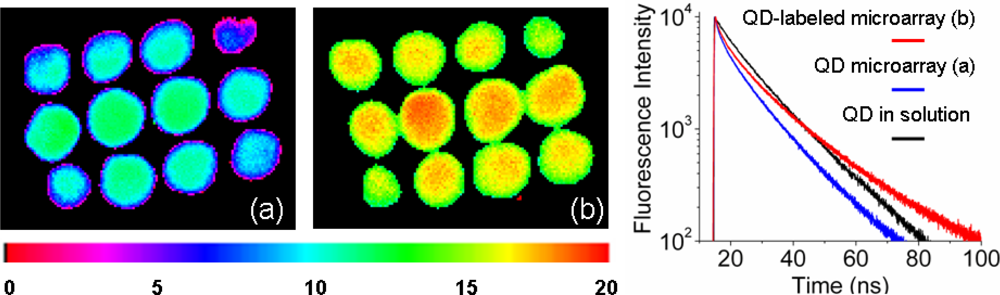
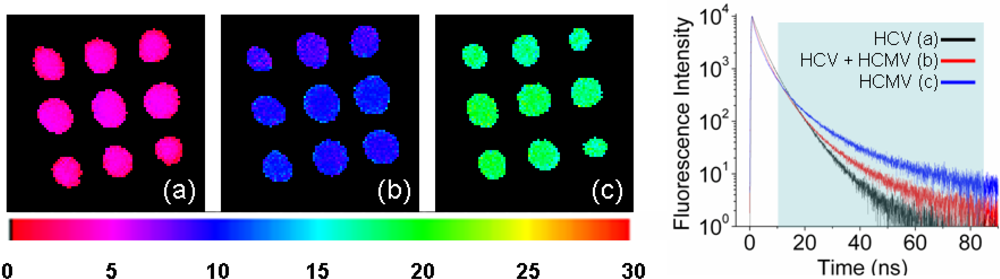
3.1. Fluorescence Lifetime Imaging of Quantum Dots and Quantum Dot-Labeled DNA Microarray spots
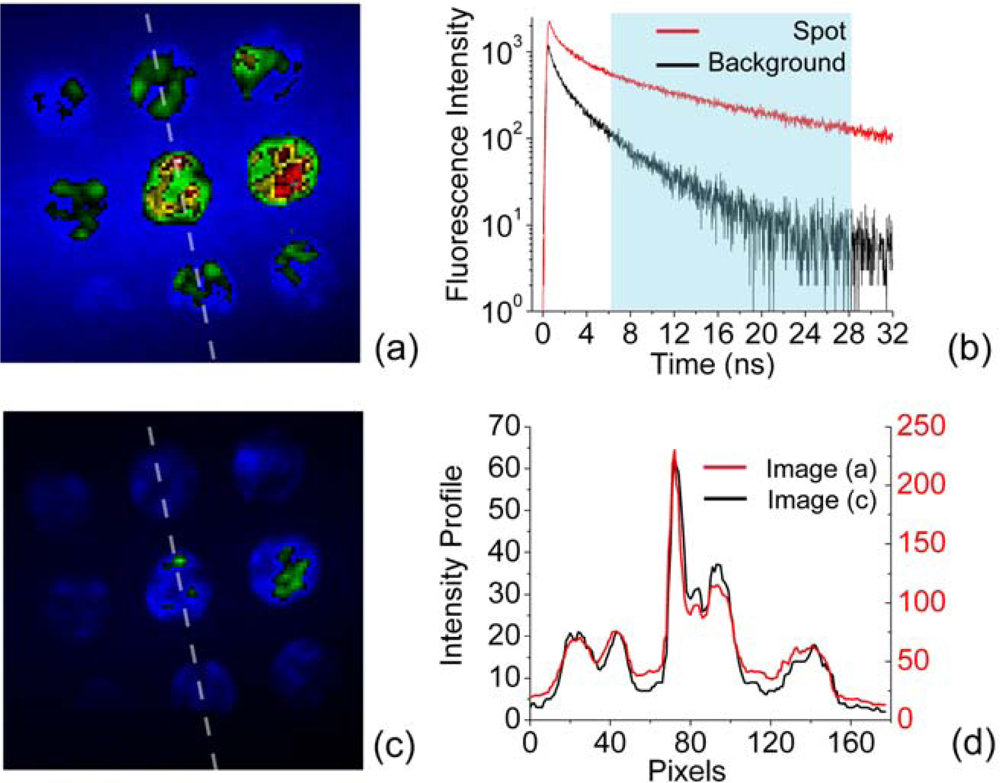
3.2. Background reduction
3.3. Multiplexed Label Detection
4. Conclusions
Acknowledgments
References
- Petrik, J. Diagnostic applications of microarrays. Transfusion Med 2006, 16, 233–247. [Google Scholar]
- Livingston, AD; Campbell, CJ; Wagner, EK; Ghazal, P. Biochip sensors for the rapid and sensitive detection of viral disease. Genome Biol 2005, 6. [Google Scholar]
- Barl, T; Dobrindt, U; Yu, XL; Katcoff, DJ; Sompolinsky, D; Bonacorsi, S; Hacker, J; Bachmann, TT. Genotyping DNA chip for the simultaneous assessment of antibiotic resistance and pathogenic potential of extraintestinal pathogenic Escherichia coli. Int. J. Antimicrob. Agents 2008, 32, 272–277. [Google Scholar]
- Situma, C; Hashimoto, M; Soper, SA. Merging microfluidics with microarray-based bioassays. Biomol. Eng 2006, 23, 213–231. [Google Scholar]
- Schaferling, M; Nagl, S. Optical technologies for the read out and quality control of DNA and protein microarrays. Anal. Bioanal. Chem 2006, 385, 500–517. [Google Scholar]
- Nagl, S; Schaeferling, M; Wolfbeis, OS. Fluorescence analysis in microarray technology. Microchim. Acta 2005, 151, 1–21. [Google Scholar]
- Woo, Y; Krueger, W; Kaur, A; Churchill, G. Experimental design for three-color and four-color gene expression microarrays. Bioinformatics 2005, 21, I459–I467. [Google Scholar]
- Lindroos, K; Sigurdsson, S; Johansson, K; Ronnblom, L; Syvanen, AC. Multiplex SNP genotyping in pooled DNA samples by a four-colour microarray system. Nucleic Acids Res 2002, 30, e70:1–e70:9. [Google Scholar]
- Shalon, D; Smith, SJ; Brown, PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res 1996, 6, 639–645. [Google Scholar]
- Suhling, K; French, PMW; Phillips, D. Time-resolved fluorescence microscopy. Photoch. Photobio. Sci 2005, 4, 13–22. [Google Scholar]
- Resch-Genger, U; Grabolle, M; Cavaliere-Jaricot, S; Nitschke, R; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar]
- Medintz, IL; Uyeda, HT; Goldman, ER; Mattoussi, H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater 2005, 4, 435–446. [Google Scholar]
- Costa-Fernandez, JM; Pereiro, R; Sanz-Medel, A. The use of luminescent quantum dots for optical sensing. Trac-Trend Anal. Chem 2006, 25, 207–218. [Google Scholar]
- Liu, TC; Liu, BS; Zhang, HL; Wang, Y. The fluorescence bioassay platforms on quantum dots nanoparticles. J. Fluoresc 2005, 15, 729–733. [Google Scholar]
- Sapsford, KE; Pons, T; Medintz, IL; Mattoussi, H. Biosensing with luminescent semiconductor quantum dots. Sensors 2006, 6, 925–953. [Google Scholar]
- Gill, R; Zayats, M; Willner, I. Semiconductor quantum dots for bioanalysis. Angew. Chem. Int. Edit 2008, 47, 7602–7625. [Google Scholar]
- Kerman, K; Endo, T; Tsukamoto, M; Chikae, M; Takamura, Y; Tamiya, E. Quantum dot-based immunosensor for the detection of prostate-specific antigen using fluorescence microscopy. Talanta 2007, 71, 1494–1499. [Google Scholar]
- Shingyoji, M; Gerion, D; Pinkel, D; Gray, JW; Chen, FQ. Quantum dots-based reverse phase protein microarray. Talanta 2005, 67, 472–478. [Google Scholar]
- Liang, RQ; Li, W; Li, Y; Tan, CY; Li, JX; Jin, YX; Ruan, KC. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res 2005, 33, e17:1–e17:8. [Google Scholar]
- Valentini, G; D’Andrea, C; Comelli, D; Pifferi, A; Taroni, P; Torricelli, A; Cubeddu, R; Battaglia, C; Consolandi, C; Salani, G; Rossi-Bernardi, L; De Bellis, G. Time-resolved DNA-microarray reading by an intensified CCD for ultimate sensitivity. Opt. Lett 2000, 25, 1648–1650. [Google Scholar]
- Waddell, E; Wang, Y; Stryjewski, W; McWhorter, S; Henry, AC; Evans, D; McCarley, RL; Soper, SA. High-resolution near-infrared imaging of DNA microarrays with time-resolved acquisition of fluorescence lifetimes. Anal. Chem 2000, 72, 5907–5917. [Google Scholar]
- Chambers, J; Angulo, A; Amaratunga, D; Guo, HQ; Jiang, Y; Wan, JS; Bittner, A; Frueh, K; Jackson, MR; Peterson, PA; Erlander, MG; Ghazal, P. DNA microarrays of the complex human cytomegalovirus genome: Profiling kinetic class with drug sensitivity of viral gene expression. J. Virol 1999, 73, 5757–5766. [Google Scholar]
- Ember, SWJ; Ross, AJ; Schulze, H; Luby, J; Bachmann, TT; Crain, J; Walton, AJ; Ciani, I; Giraud, G; Mount, AR; Tlili, C; Terry, JG; Khondoker, MR; Ghazal, P; Campbell, CJ. Clinical applications of DNA and protein microarrays for the serodiagnosis of HCV infection To be submitted for publication.
- Emiliani, V; Sanvitto, D; Tramier, M; Piolot, T; Petrasek, Z; Kemnitz, K; Durieux, C; Coppey-Moisan, M. Low-intensity two-dimensional imaging of fluorescence lifetimes in living cells. Appl. Phys. Lett 2003, 83, 2471–2473. [Google Scholar]
- Grecco, HE; Lidke, KA; Heintzmann, R; Lidke, DS; Spagnuolo, C; Martinez, OE; Jares-Erijman, EA; Jovin, TM. Ensemble and single particle photophysical proper-ties (Two-Photon excitation, anisotropy, FRET, lifetime, spectral conversion) of commercial quantum dots in solution and in live cells. Microsc. Res. Techniq 2004, 65, 169–179. [Google Scholar]
- Fisher, BR; Eisler, HJ; Stott, NE; Bawendi, MG. Emission intensity dependence and single-exponential behavior in single colloidal quantum dot fluorescence lifetimes. J. Phys. Chem. B 2004, 108, 143–148. [Google Scholar]
- Jones, M; Nedeljkovic, J; Ellingson, RJ; Nozik, AJ; Rumbles, G. Photoenhancement of luminescence in colloidal CdSe quantum dot solutions. J. Phys. Chem. B 2003, 107, 11346–11352. [Google Scholar]
- Kloepfer, JA; Bradforth, SE; Nadeau, JL. Photophysical properties of biologically compatible CdSe quantum dot structures. J. Phys. Chem. B 2005, 109, 9996–10003. [Google Scholar]
- Algar, WR; Krull, UJ. Luminescence and stability of aqueous thioalkyl acid capped CdSe/ZnS quantum dots correlated to ligand ionization. ChemPhysChem 2007, 8, 561–568. [Google Scholar]
- Sutter, JU; Macmillan, AM; Birch, DJS; Rolinski, OJ. Toward single-metal-ion sensing by Forster resonance energy transfer. In Annals of the New York Academy of Sciences, 1st Ed; Wolfbeis, OS, Ed.; Blackwell Publishing: Oxford, UK, 2008; Volume 1130, pp. 62–67. [Google Scholar]
- Thomas, AV; Herl, L; Spoelgen, R; Hiltunen, M; Jones, PB; Tanzi, RE; Hyman, BT; Berezovska, O. Interaction between presenilin 1 and ubiquilin 1 as detected by fluorescence lifetime imaging microscopy and a high-throughput fluorescent plate reader. J. Biol. Chem 2006, 281, 26400–26407. [Google Scholar]
- Niclass, C; Favi, C; Kluter, T; Gersbach, M; Charbon, E. A 128 × 128 Single-Photon Image Sensor With Column-Level 10-Bit Time-to-Digital Converter Array. IEEE J. Solid-St. Circ 2008, 43, 2977–2989. [Google Scholar]
- Charbon, E. Towards large scale CMOS single-photon detector arrays for lab-on-chip applications. J Phys D: Appl Phys 2008, 41. [Google Scholar]
| HCMV target | 5′-AGTGTTGAGGGCCGTAAGCGTGTTGTGTCCGACGCTGCCTGCGCA
CTGCCGGTGCGTGTCGTCCCACGGTATTTG – 3′ [5′] = Biotin-TEG |
| HCMV probe | 5′-CAAATACCGTGGGACGACACGCACCGGCAGTGCGCAGGCAGCGT
CGGACACAACACGCTTACGGCCCTCAACACT – 3′ [5′] = SH (C6) |
| HCV target | 5′-GCGAAGGCTTGTGGTACTGCCTGATAGGGTGCTTGCGAGTGCCCC
GGGAGGTCTCGTAGACCGTGCA [5′] = Alexa 430 |
| HCV probe | 5′-TGCACGGTCTACGAGACCTCCCGGGGCACTCGCAAGCACCCTATC
AGGCAGTACCACAAGGCCTTTCGC - 3′ [5′] = SH |
| Sample | Free QD Solution* | Free QD Glass | QD-dsDNA Glass |
|---|---|---|---|
| t1 (ns) | 2.1 ± 0.05 | 1.6 ± 0.02 | 1.9 ± 0.02 |
| t2 (ns) | 12.1 ± 0.1 | 9.6 ± 0.05 | 11.1 ± 0.1 |
| t3 (ns) | 27.6 ± 0.3 | 21.8 ± 0.2 | 29.5 ± 0.2 |
| A1 (%) | 27.2 ± 0.4 | 32.4 ± 0.2 | 30 ± 0.2 |
| A2 (%) | 63.5 ± 0.3 | 54.1 ± 0.2 | 52.6 ± 0.2 |
| A3 (%) | 9.4 ± 0.4 | 13.4 ± 0.3 | 17.4 ± 0.3 |
| χ2 | 1.3 | 1.5 | 1.4 |
| Tc (ns) | 15.3 | 13.3 | 18.8 |
| Spot | QD-labeled HCMV spot | Alexa-labeled HCV spot | Mixed spot HCMV/HCV |
|---|---|---|---|
| t1 (ns) | 0.67 ± 0.01 | 1.4 ± 0.02 | 0.48 ± 0.05 |
| t2 (ns) | 2.5 ± 0.03 | 3.63 ± 0.03 | 2.1 ± 0.1 |
| t3 (ns) | 6.8 ± 0.1 | 7.5 ± 0.1 | 5 ± 0.3 |
| t4 (ns) | 23.9 ± 0.3 | NA | 16.8 ± 0.3 |
| A1 (%) | 36.1 ± 0.4 | 27.1 ± 0.7 | 20.6 ± 0.3 |
| A2 (%) | 48 ± 0.3 | 63.5 ± 0.5 | 47.2 ± 0.3 |
| A3 (%) | 14.7 ± 0.4 | 9.4 ± 0.5 | 31 ± 0.5 |
| A4 (%) | 1.25 ± 0.04 | NA | 1.25 ± 0.03 |
| χ2 | 2.4 | 1.5 | 2.9 |
| Tc (ns) | NA | 4.2 | NA |
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Giraud, G.; Schulze, H.; Bachmann, T.T.; Campbell, C.J.; Mount, A.R.; Ghazal, P.; Khondoker, M.R.; Ross, A.J.; Ember, S.W.J.; Ciani, I.; et al. Fluorescence Lifetime Imaging of Quantum Dot Labeled DNA Microarrays. Int. J. Mol. Sci. 2009, 10, 1930-1941. https://doi.org/10.3390/ijms10041930
Giraud G, Schulze H, Bachmann TT, Campbell CJ, Mount AR, Ghazal P, Khondoker MR, Ross AJ, Ember SWJ, Ciani I, et al. Fluorescence Lifetime Imaging of Quantum Dot Labeled DNA Microarrays. International Journal of Molecular Sciences. 2009; 10(4):1930-1941. https://doi.org/10.3390/ijms10041930
Chicago/Turabian StyleGiraud, Gerard, Holger Schulze, Till T. Bachmann, Colin J. Campbell, Andrew R. Mount, Peter Ghazal, Mizanur R. Khondoker, Alan J. Ross, Stuart W. J. Ember, Ilenia Ciani, and et al. 2009. "Fluorescence Lifetime Imaging of Quantum Dot Labeled DNA Microarrays" International Journal of Molecular Sciences 10, no. 4: 1930-1941. https://doi.org/10.3390/ijms10041930
APA StyleGiraud, G., Schulze, H., Bachmann, T. T., Campbell, C. J., Mount, A. R., Ghazal, P., Khondoker, M. R., Ross, A. J., Ember, S. W. J., Ciani, I., Tlili, C., Walton, A. J., Terry, J. G., & Crain, J. (2009). Fluorescence Lifetime Imaging of Quantum Dot Labeled DNA Microarrays. International Journal of Molecular Sciences, 10(4), 1930-1941. https://doi.org/10.3390/ijms10041930