Isolation and Characterization of Novel Microsatellite Markers for Yellow Perch (Perca flavescens)
Abstract
:1. Introduction
2. Experimental Section
2.1. EST database mining
2.2. Microsatellite-enriched library construction
2.3. Sequence analysis and primer design
2.4. DNA extraction, PCR amplification and genotyping
2.5. Genetic data analysis
2.6. Cross utility
3. Results and Discussion
3.1. Genomic-SSRs
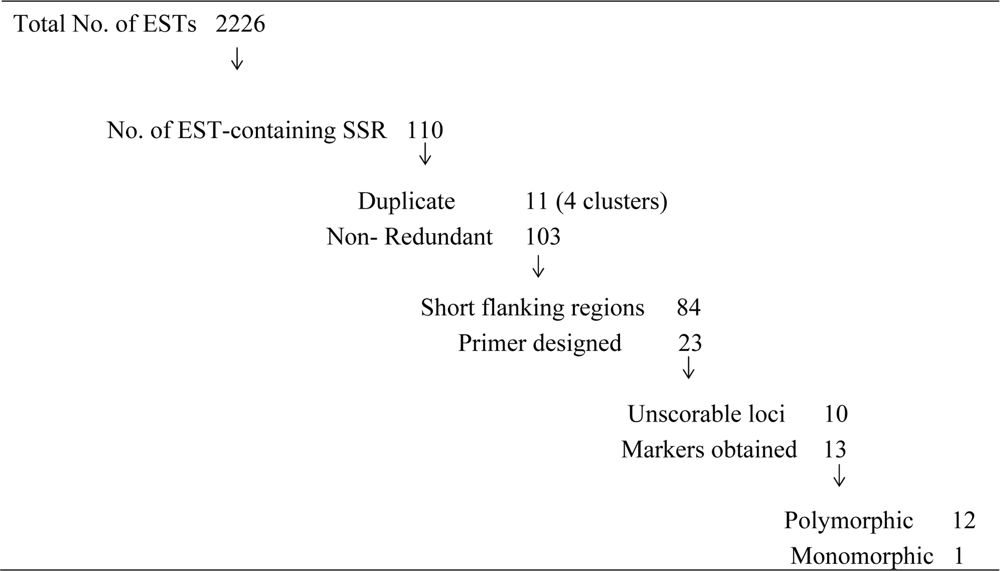
3.2. EST-SSRs
3.3. Cross utility
4. Conclusions
Acknowledgments
References
- Malison, JA. A white paper on the status and needs of yellow perch aquaculture in the North Carolina regions; North Central Regional Aquaculture Center, Michigan State University: East Lansing, Michigan, USA.
- Leclerc, D; Wirth, T; Bernatchez, L. Isolation and characterization of microsatellite loci in yellow perch (Perca flavescens), and cross-species amplification within the family Percidae. Mol. Ecol. Notes 2000, 9, 995–997. [Google Scholar]
- Li, L; Wang, HP; Givens, C; Czesny, S; Brown, B. Isolation and characterization of microsatellites in yellow perch (Perca flavescens). Mol. Ecol. Notes 2007, 7, 600–603. [Google Scholar]
- Serapion, J; Kucuktas, H; Feng, J; Liu, Z. Bioinformatic mining of type I microsatellites from expressed sequence tags of Channel catfish (Ictalurus punctatus). Mar. Biotechnol 2004, 6, 364–377. [Google Scholar]
- Zhan, AB; Bao, ZM; Wang, XL; Hu, JJ. Microsatellite markers derived from bay scallop Argopecten irradians expressed sequence tags. Fish. Sci 2005, 71, 1341–1346. [Google Scholar]
- Zhan, AB; Hu, JJ; Hu, XL; Wang, ML; Peng, W; Li, Y; Bao, ZM. Isolation and characterization of microsatellite markers for Zhikong scallop by screening SSR-enriched library. J. Fish. China 2008, 32, 353–361, (in Chinese with English abstract). [Google Scholar]
- Wang, D; Liao, X; Cheng, L; Yu, X; Tong, J. Development of novel EST-SSR markers in common carp by data mining from public EST sequences. Aquaculture 2007, 271, 558–574. [Google Scholar]
- Zhou, ZC; Zou, LL; Dong, Y; He, CB; Liu, WD; Deng, H; Wang, LM. Characterization of 28 polymorphic microsatellites for Japanese sea urchin (Strongylocentrotus intermedius) via mining EST database of a related species (S. purpuratus). Ann. Zool. Fenn 2008, 45, 181–184. [Google Scholar]
- Boutin-Ganache, I; Raposo, M; Raymond, M. M13-tailed primers improve the readability and usability of microsatellite analysis performed with two different allele sizing methods. BioTechniques 2001, 31, 24–28. [Google Scholar]
- Waters, J; Epifanio, J; Gunter, T; Brown, B. Homing behaviour facilitates subtle population differentiation among river populations of Alosa sapidissima: Microsatellites and mtDNA. J. Fish Bio 2000, 56, 622–636. [Google Scholar]
- Guo, SW; Thompson, EA. Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics 1992, 48, 361–372. [Google Scholar]
- Rice, WR. Analyzing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar]
- Van Oosterhout, C; Hutchinson, WF; Wills, DPM; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar]
- Brown, B; Wang, HP; Li, L; Givens, C; Wallat, G. Yellow perch strain evaluation I: Genetic variation of six brood stock populations. Aquaculture 2007, 271, 142–151. [Google Scholar]
- Leclerc, E; Mailhot, Y; Mingelbier, M; Bernatchez, L. The landscape genetics of yellow perch (Perca flavescens) in a large fluvial ecosystem. Mol. Ecol 2008, 17, 1702–1717. [Google Scholar]
- Wang, Y; Guo, X. Development and characterization of EST-SSR markers in the eastern oyster Crassostrea virginica. Mar. Biotechnol 2008, 9, 500–511. [Google Scholar]
- Edwards, YJ; Elgar, G; Clark, MS; Bishop, MJ. The identification and haracterization of microsatellites in the compact genome of the Japanese pufferfish, Fugu rubripes: erspectives in functional and comparative genomic analyses. J. Mol. Biol 1998, 278, 843–854. [Google Scholar]
- Serapion, J; Kucuktas, H; Feng, JN; Liu, ZJ. Bioinformatic mining of type I microsatellites from expressed sequence tags of channel catfish (Ictalurus punctatus). Mar. Biotechnol 2004, 6, 364–377. [Google Scholar]
- Zhang, LL; Chen, C; Cheng, J; Wang, S; Hu, XL; Hu, JJ; Bao, ZM. Initial analysis of tandemly repetitive sequences in the genome of Zhikong scallop (Chlamys farreri Jones et Preston). DNA sequence 2007, 19, 195–205. [Google Scholar]
- Zhan, AB; Bao, ZM; Hu, XL; Wang, S; Peng, W; Wang, ML; Hu, JJ; Liang, CZ; Yue, ZQ. Characterization of 95 novel microsatellite markers for Zhikong scallop Chlamys farreri using FIASCO-colony hybridization and EST database mining. Fish. Sci 2008, 74, 516–526. [Google Scholar]
- Eujayl, I; Sorrells, ME; Baum, M; Wolters, P; Powell, W. Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor. Appl. Genet 2002, 104, 399–407. [Google Scholar]
| Locus name Accession | Primer sequence (5′-3′) | Ta | Repeats | S (bp) | A | Ho | He | P-value | Cross utility (Ta; A) |
|---|---|---|---|---|---|---|---|---|---|
| YP23† FJ547096 | F: M13-TTGGACAAAAATAACTCACT
R: AGAGTAGAAATGCGGTTGCT | 55 | (TTC)16 | 180–210 | 10 | 0.8077 | 0.8620 | 0.8312 | 52; 3 |
| YP72† FJ547097 | F: AAAGAGAGCAAAGGGGAAGA
R: M13-TGTGTAAGAAACAGGCAGGT | 55 | (GGT)5GAA (GGT)5GAA(GGT)16 | 255–264 | 3 | 0.3846 | 0.4970 | 0.4615 | 54; 2 |
| YP86† FJ547098 | F: M13-CCGGCTACTTCATGTTAAAA
R: GTGGGAATAAGGGTTAGGCT | 55 | (AGAT)14 | 331–387 | 12 | 0.5185 | 0.9371 | 0.0093* | — |
| YP89† FJ547099 | F: ATGGAGATTTACAGCCCCTA
R: M13-ACTAATAACCACCATCCTGC | 55 | (CA)5GA(CA)18 | 191–227 | 6 | 0.1238 | 0.6260 | 0.0000* | — |
| YP90† FJ547100 | F: M13-AGAAAAGAGGGAAAGAAGG
R: CCGCTATTTCACTCTGTTTT | 52 | (GAAA)16 | 123–171 | 11 | 0.5556 | 0.7596 | 0.8084 | — |
| YP94† FJ547101 | F: M13-TTCACATTCAATAGGAGTAGAGT
R: CTGTAAAACCATTGCCGATAAA | 50 | (ACAT)15 | 331–407 | 9 | 0.0714 | 0.8331 | 0.0003* | — |
| YP95† FJ547102 | F: GTGCCCTTTGTCACCCAT
R: M13-GCCCTCATTTATGTCTCTCC | 55 | (CA)14 | 127–133 | 3 | 0.0870 | 0.3710 | 0.0001* | 52; 1 |
| YP105† FJ547103 | F: M13-TAGAAGCAAAACCCGTGA
R: TGTCCCTCACCAGCCAGT | 55 | (CTA)14 | 169–214 | 14 | 0.4815 | 0.9511 | 0.0028* | — |
| PFE01# DR730576 | F: M13-CTCCCAAAATAAAGCCAATGTC
R: ACAGAGTTTCAGGCACTTGTGG | 54 | (TC)10 | 250–268 | 2 | 0.0714 | 0.0701 | 0.8907 | 54; 2 |
| PFE03# DR730639 | F: M13-GCAGAAATGCTACATAGATCCT
R: AGTCAATATCCTCCAAATGTGC | 52 | (GT)16 | 124–136 | 5 | 0.5714 | 0.5396 | 0.8719 | 50; 3 |
| PFE06#
DV671343 | F: M13-TTGCCTGAGGTTGTATTGAGAA
R: ACAGTCGTAGCAGAGGGTCAC | 52 | (AG)7 | 164–176 | 2 | 0.0357 | 0.0357 | 1.0000 | 52; 2 |
| PFE07# DV671312 | F: M13-CGGCACGAGGGGACTGTAATC
R: TGTGCTCTTTCCCTTGTGACCG | 50 | (AAC)6 | 109–121 | 3 | 0.0357 | 0.1045 | 0.0018* | 54; 1 |
| PFE08# DV671070 | F: M13-GTCTTAAACAAGTCTTCATAGCAC
R: GGACAGAGAACACATAGAGAATC | 56 | (TAA)11 | 160–168 | 2 | 0.0357 | 0.0357 | 1.0000 | 50; 1 |
| PFE11# DW985750 | F: M13-CTTAGACAGACCGACCTACAG
R: ATGTCAGCCAAGATGTAATG | 50 | (TGA)12 | 220–223 | 2 | 0.0357 | 0.0357 | 1.0000 | — |
| PFE12# DV752650 | F: M13-TGCGTGCCAAGGGCGGTGTT
R: CCGTCCCCTCAACAAATACC | 54 | (CCT)5 | 131–149 | 3 | 0.0357 | 0.0708 | 0.0018* | 54; 1 |
| PFE14# DV671188 | F: M13-AGCCACAAAGCTGAACATAG
R: TGCCATGTTGTATCTCCCAC | 52 | (AT)10 | 258–264 | 3 | 0.1429 | 0.1351 | 0.7270 | 50; 1 |
| PFE15# DR731110 | F: M13-GTATTAGTCTATGTATATTGCC
R: CGGGATGTCACTTACTTCTC | 55 | (TATC)17 | 292–296 | 2 | 0.0357 | 0.0357 | 1.0000 | 50; 1 |
| PFE19# DV671307 | F: M13-TGTCTAACGATTGCTTTTCCT
R: CAATGAAAAATAAACATGCGTGACC | 56 | (AT)10 | 80–82 | 2 | 0.0000 | 0.0701 | 0.0016* | — |
| PFE20# DR731052 | F: M13-GATCCATCCTGCTCAGACTC
R: AAGAGATTGAGTTTGGTAGC | 56 | (TC)23 | 281–283 | 2 | 0.0000 | 0.0701 | 0.0016* | — |
| PFE22# DR730585 | F: M13-ATACAGAGGCCTTCATTTGT
R: CAGCTACAGTTCATTCTACCT | 56 | (TA)9 | 280–282 | 3 | 0.0714 | 0.0701 | 0.8907 | — |
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/). This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhan, A.; Wang, Y.; Brown, B.; Wang, H.-P. Isolation and Characterization of Novel Microsatellite Markers for Yellow Perch (Perca flavescens). Int. J. Mol. Sci. 2009, 10, 18-27. https://doi.org/10.3390/ijms10010018
Zhan A, Wang Y, Brown B, Wang H-P. Isolation and Characterization of Novel Microsatellite Markers for Yellow Perch (Perca flavescens). International Journal of Molecular Sciences. 2009; 10(1):18-27. https://doi.org/10.3390/ijms10010018
Chicago/Turabian StyleZhan, Aibin, Yao Wang, Bonnie Brown, and Han-Ping Wang. 2009. "Isolation and Characterization of Novel Microsatellite Markers for Yellow Perch (Perca flavescens)" International Journal of Molecular Sciences 10, no. 1: 18-27. https://doi.org/10.3390/ijms10010018




