Production of a Novel Tetrahydroxynaphthalene (THN) Derivative from Nocardia sp. CS682 by Metabolic Engineering and Its Bioactivities
Abstract
:1. Introduction
2. Results and Discussion
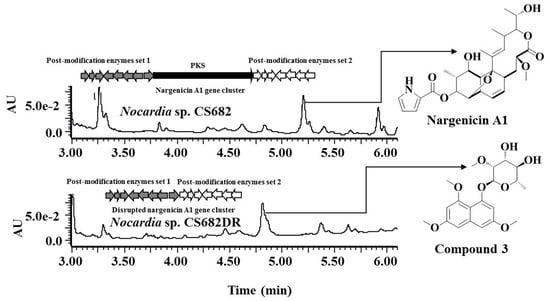
2.1. Construction of Nargenicin Deletion Mutant
2.2. Fermentation, Isolation, and Structural Elucidation of the Compound IBR-3 from Nocardia sp. CS682DR
2.3. Metabolite Profiling and Mass Spectrometric Analysis
2.4. Bioinformatic Analysis of Novel THN Gene Cluster and Proposed Biosynthetic Pathway
2.5. Analysis of Acyl-CoAs and Transcript Level
2.6. Bioactivities of Novel THN Derivative
3. Materials and Methods
3.1. General Information
3.2. Inactivation of Nargenicin A1 Biosynthetic Gene Cluster and Comparative Analysis
3.3. Fermentation and Isolation of Compound IBR-3
3.4. Bioinformatic Analysis
3.5. Extraction and Analysis of Acyl-CoAs and Transcript Level
3.6. Evaluation of Biological Activities
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dhakal, D.; Sohng, J.K. Coalition of biology and chemistry for ameliorating antimicrobial drug discovery. Front. Microbiol. 2017, 8, 734. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, D.; Pokhrel, A.R.; Shrestha, B.; Sohng, J.K. Marine rare actinobacteria: Isolation, characterization, and strategies for harnessing bioactive compounds. Front. Microbiol. 2017, 8, 1106. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.; Reker, D.; Schneider, P.; Schneider, G. Counting on natural products for drug design. Nat. Chem. 2016, 8, 531. [Google Scholar] [CrossRef] [PubMed]
- Behal, V. Bioactive products from Streptomyces. Adv. Appl. Microbiol. 2000, 47, 113–156. [Google Scholar]
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Jebakumar, S.R. Non-streptomycete actinomycetes nourish the current microbial antibiotic drug discovery. Front. Microbiol. 2013, 4, 240. [Google Scholar] [CrossRef]
- Dhakal, D.; Sohng, J.K. Laboratory maintenance of Nocardia species. Curr. Protoc. Microbiol. 2015, 39. [Google Scholar] [CrossRef]
- Luo, Q.; Hiessl, S.; Steinbüchel, A. Functional diversity of Nocardia in metabolism. Environ. Microbiol. 2014, 16, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, M.; Hayashi, C.; Homma, Y.; Hattori, S.; Kinoshita, N.; Hamada, M.; Takeuchi, T. Tubelactomicin A, a Novel 16-Membered Lactone Antibiotic, from Nocardia sp. J. Antibiot. 2000, 53, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Sohng, J.K.; Yamaguchi, T.; Seong, C.N.; Baik, K.S.; Park, S.C.; Lee, H.J.; Jang, S.Y.; Simkhada, J.R.; Yoo, J.C. Production, isolation and biological activity of nargenicin from Nocardia sp. CS682. Arch. Pharm. Res. 2008, 31, 1339–1345. [Google Scholar] [CrossRef]
- Tsuda, M.; Sato, H.; Tanaka, Y.; Yazawa, K.; Mikami, Y.; Sasaki, T.; Kobayashi, J.I. Brasiliquinones A–C, new cytotoxic benz [a] anthraquinones with an ethyl group at C-3 from actinomycete Nocardia brasiliensis. J. Chem. Soc. Perkin. 1996, 15, 1773–1775. [Google Scholar] [CrossRef]
- Shigemori, H.; Komaki, H.; Yazawa, K.; Mikami, Y.; Nemoto, A.; Tanaka, Y.; Sasaki, T.; In, Y.; Ishida, T.; Kobayashi, J.I.; et al. A Novel Tricyclic Metabolite with Potent Immunosuppressive Activity from Actinomycete Nocardia brasiliensis. J. Org. Chem. 1998, 63, 6900–6904. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhang, L.; Zhou, Z.; Yan, X. Diversity of gene clusters for polyketide and nonribosomal peptide biosynthesis revealed by metagenomic analysis of the yellow sea sediment. Front. Microbiol. 2018, 9, 295. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Du, L. Iterative polyketide biosynthesis by modular polyketide synthases in bacteria. Appl. Microbiol. Biotechnol. 2016, 100, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Ridley, C.P.; Lee, H.Y.; Khosla, C. Evolution of polyketide synthases in bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 4595–4600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 2003, 7, 285–295. [Google Scholar] [CrossRef]
- Austin, M.B.; Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2003, 20, 79–110. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Xu, F.; Zeng, J.; Zhan, J. Type III polyketide synthases in natural product biosynthesis. IUBMB Life 2012, 64, 285–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, Y.; Ogata, H.; Goto, S. Type III polyketide synthases: Functional classification and phylogenomics. ChemBioChem 2017, 18, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Cortés, J.; Velasco, J.; Foster, G.; Blackaby, A.P.; Rudd, B.A.; Wilkinson, B. Identification and cloning of a type III polyketide synthase required for diffusible pigment biosynthesis in Saccharopolyspora erythraea. Mol. Microbiol. 2002, 44, 1213–1224. [Google Scholar] [CrossRef]
- Funa, N.; Ohnishi, Y.; Ebizuka, Y.; Horinouchi, S. Properties and substrate specificity of RppA, a chalcone synthase-related polyketide synthase in Streptomyces griseus. J. Biol. Chem. 2002, 277, 4628–4635. [Google Scholar] [CrossRef] [PubMed]
- Izumikawa, M.; Shipley, P.R.; Hopke, J.N.; Thomas, O.; Xiang, L.; Noel, J.P.; Moore, B.S. Expression and characterization of the type III polyketide synthase 1, 3, 6, 8-tetrahydroxynaphthalene synthase from Streptomyces coelicolor A3 (2). J. Ind. Microbiol. Biotechnol. 2003, 30, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, G.P.; Oh, T.J.; Liou, K.; Sohng, J.K. Identification of a cryptic type III polyketide synthase (1, 3, 6, 8-tetrahydroxynaphthalene synthase) from Streptomyces peucetius ATCC 27952. Mol. Cells 2008, 26, 362–367. [Google Scholar] [PubMed]
- Zeng, J.; Decker, R.; Zhan, J. Biochemical characterization of a type III polyketide biosynthetic gene cluster from Streptoycems toxytricini. Appl. Biochem. Biotechnol. 2012, 166, 1020–1033. [Google Scholar] [CrossRef]
- Gross, F.; Luniak, N.; Perlova, O.; Gaitatzis, N.; Jenke-Kodama, H.; Gerth, K.; Gottschalk, D.; Dittmann, E.; Müller, R. Bacterial type III polyketide synthases: Phylogenetic analysis and potential for the production of novel secondary metabolites by heterologous expression in pseudomonads. Arch. Microbiol. 2006, 185, 28–38. [Google Scholar] [CrossRef]
- Zhao, B.; Guengerich, F.P.; Bellamine, A.; Lamb, D.C.; Izumikawa, M.; Lei, L.; Podust, L.M.; Sundaramoorthy, M.; Kalaitzis, J.A.; Reddy, L.M.; et al. Binding of two flaviolin substrate molecules, oxidative coupling, and crystal structure of Streptomyces coelicolor A3 (2) cytochrome P450 158A2. J. Biol. Chem. 2005, 280, 11599–11607. [Google Scholar] [CrossRef] [PubMed]
- Kuzuyama, T.; Noel, J.P.; Richard, S.B. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 2005, 435, 983–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhakal, D.; Lim, S.K.; Kim, D.H.; Kim, B.G.; Yamaguchi, T.; Sohng, J.K. Complete genome sequence of Streptomyces peucetius ATCC 27952, the producer of anticancer anthracyclines and diverse secondary metabolites. J. Biotechnol. 2018, 267, 50–54. [Google Scholar] [CrossRef]
- Thuan, N.H.; Dhakal, D.; Pokhrel, A.R.; Chu, L.L.; Van Pham, T.T.; Shrestha, A.; Sohng, J.K. Genome-guided exploration of metabolic features of Streptomyces peucetius ATCC 27952: Past, current, and prospect. Appl. Microbiol. Biotechnol. 2018, 102, 4355–4370. [Google Scholar] [CrossRef]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes–a review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef]
- Baltz, R.H. Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L. Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology 2008, 154, 1555–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhang, C.; Liu, C.; Ju, J.; Ma, J. Genome sequencing of Streptomyces atratus SCSIOZH16 and activation production of Nocardamine via metabolic engineering. Front. Microbiol. 2018, 13, 1269. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Oh, T.J.; Sohng, J.K. Exploration of geosmin synthase from Streptomyces peucetius ATCC 27952 by deletion of doxorubicin biosynthetic gene cluster. J. Ind. Microbiol. Biotechnol. 2009, 36, 1257–1265. [Google Scholar] [CrossRef]
- Dhakal, D.; Chaudhary, A.K.; Yi, J.S.; Pokhrel, A.R.; Shrestha, B.; Parajuli, P.; Shrestha, A.; Yamaguchi, T.; Jung, H.J.; Kim, S.Y.; et al. Enhanced production of nargenicin A1 and creation of a novel derivative using a synthetic biology platform. Appl. Microbiol. Biotechnol. 2016, 100, 9917–9931. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, D.; Le, T.T.; Pandey, R.P.; Jha, A.K.; Gurung, R.; Parajuli, P.; Pokhrel, A.R.; Yoo, J.C.; Sohng, J.K. Enhanced production of nargenicin A 1 and generation of novel glycosylated derivatives. Appl. Biochem. Biotechnol. 2015, 175, 2934–2949. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Ramos, A.; Lombo, F.; Brana, A.F.; Rohr, J.; Mendez, C.; Salas, J.A. Biosynthesis of elloramycin in Streptomyces olivaceus requires glycosylation by enzymes encoded outside the aglycon cluster. Microbiology 2008, 154, 781–788. [Google Scholar] [CrossRef]
- Madduri, K.; Waldron, C.; Merlo, D.J. Rhamnose biosynthesis pathway supplies precursors for primary and secondary metabolism in Saccharopolyspora spinosa. J. Bacteriol. 2001, 183, 5632–5638. [Google Scholar] [CrossRef]
- Singh, B.; Lee, C.B.; Sohng, J.K. Precursor for biosynthesis of sugar moiety of doxorubicin depends on rhamnose biosynthetic pathway in Streptomyces peucetius ATCC 27952. Appl. Microbiol. Biotechnol. 2010, 85, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Kaysser, L.; Wemakor, E.; Siebenberg, S.; Salas, J.A.; Sohng, J.K.; Kammerer, B.; Gust, B. Formation and attachment of the deoxysugar moiety and assembly of the gene cluster for caprazamycin biosynthesis. Appl. Environ. Microbiol. 2010, 76, 4008–4018. [Google Scholar] [CrossRef] [PubMed]
- Cairns, T.; Meyer, V. In silico prediction and characterization of secondary metabolite biosynthetic gene clusters in the wheat pathogen Zymoseptoria tritici. BMC Genom. 2017, 18, 631. [Google Scholar] [CrossRef] [PubMed]
- Cane, D.E.; Yang, C.C. Biosynthetic origin of the carbon skeleton and oxygen atoms of nargenicin A1. J. Am. Chem. Soc. 1984, 106, 784–787. [Google Scholar] [CrossRef]
- Hardt, I.H.; Jensen, P.R.; Fenical, W. Neomarinone, and new cytotoxic marinone derivatives, produced by a marine filamentous bacterium (actinomycetales). Tetrahedron Lett. 2000, 41, 2073–2076. [Google Scholar] [CrossRef]
- Kawasaki, T.; Hayashi, Y.; Kuzuyama, T.; Furihata, K.; Itoh, N.; Seto, H.; Dairi, T. Biosynthesis of a natural polyketide-isoprenoid hybrid compound, furaquinocin A: Identification and heterologous expression of the gene cluster. J. Bacteriol. 2006, 188, 1236–1244. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, N.E.A.; El-Ewasy, S.M. Bioproduction, characterization, anticancer and antioxidant activities of extracellular melanin pigment produced by newly isolated microbial cell factories Streptomyces glaucescens NEAE-H. Sci Rep. 2017, 7, 42129. [Google Scholar] [CrossRef]
- Korkina, L.; Kostyuk, V.; Potapovich, A.; Mayer, W.; Talib, N.; De Luca, C. Secondary Plant Metabolites for Sun Protective Cosmetics: From Pre-Selection to Product Formulation. Cosmetics 2018, 5, 32. [Google Scholar] [CrossRef]
- Dhakal, D.; Jha, A.K.; Pokhrel, A.; Shrestha, A.; Sohng, J.K. Genetic manipulation of Nocardia species. Curr. Protoc. Microbiol. 2016, 40. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014, 1079, 105–116. [Google Scholar] [CrossRef]
- Totowa, N.J.; Page, R.D.M. TreeView. An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996, 12, 357–358. [Google Scholar]
- Pokhrel, A.R.; Nguyen, H.T.; Dhakal, D.; Chaudhary, A.K.; Sohng, J.K. Implication of orphan histidine kinase (OhkAsp) in biosynthesis of doxorubicin and daunorubicin in Streptomyces peucetius ATCC 27952. Microbiol Res. 2018, 214, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Le, T.T.; Pandey, R.P.; Gurung, R.B.; Dhakal, D.; Sohng, J.K. Efficient enzymatic systems for synthesis of novel α-mangostin glycosides exhibiting antibacterial activity against Gram-positive bacteria. Appl. Microbiol. Biotechnol. 2014, 98, 8527–8538. [Google Scholar] [CrossRef] [PubMed]
- Gurung, R.B.; Gong, S.Y.; Dhakal, D.; Le, T.T.; Jung, N.R.; Jung, H.J.; Oh, T.J.; Sohng, J.K. Synthesis of curcumin glycosides with enhanced anticancer properties using one-pot multienzyme glycosylation technique. J. Microbiol. Biotechnol. 2017, 27, 1639–1648. [Google Scholar] [CrossRef] [PubMed]
- Sim, D.Y.; Sohng, J.K.; Jung, H.J. Anticancer activity of 7, 8-dihydroxyflavone in melanoma cells via downregulation of α-MSH/cAMP/MITF pathway. Oncol. Rep. 2016, 36, 528–534. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compound IBR-3 are available from the authors. |







| Position | δc | δH (J in Hz) | Intensities |
|---|---|---|---|
| 1′ | 96.38 | 5.48 (s) | 1H |
| 2′ | 80.51 | 3.58 (m) | |
| 3′ | 70.26 | 3.88(dd) J = 9.6 Hz, 9.6 Hz, | |
| 4′ | 72.19 | 3.25(dd) J = 9.5 Hz, 9.5 Hz | |
| 5′ | 69.7 | 3.58 (m) | |
| 6′ | 17.92 | 1.10(d) J = 6.2 Hz | 3H |
| 1 | 153.92 | ||
| 2 | 102.15 | 6.56(d) J = 2.4 Hz | |
| 3 | 157.87 | ||
| 4 | 100.61 | 6.87(d) J = 2.4 Hz | |
| 5 | 98.52 | 6.78(d) J = 2.3 Hz | |
| 6 | 158.32 | ||
| 7 | 96.5 | 6.38(d) J = 2.3 Hz | |
| 8 | 157.56 | ||
| 9 | 55.09 | 3.81(s) | 3H |
| 10 | 55.08 | 3.82(s) | |
| 11 | 55.8 | 3.84(s) | |
| 12 | 58.73 | 3.44(s) | 3H |
| 4a | 138.76 | ||
| 8a | 108.62 |
| Compound | Name | Structure | Elemental formula | [M + H]+ theoretical | [M + H]+ observed | Peak in UPLC | tR (min) |
|---|---|---|---|---|---|---|---|
| IBR-1 | 3,6,8-trimethoxy naphthalen-1-ol |  | C13H14O4 | 235.0965 | 235.0961 | 4 | 5.37 |
| IBR-2 | 1-(α-l-6-deoxy-mannopyranosyloxy)- 3,6,8-trimethoxy naphthalene |  | C19H24O8 | 381.1544 | 381.1543 | 2 | 4.46 |
| IBR-3 | 1-(α-l-(2-O-methyl)-6-deoxymanno- pyranosyloxy)-3,6,8-trimethoxy naphthalene |  | C20H26O8 | 395.1700 | 395.1705 | 3 | 4.81 |
| IBR-4 | 1,3,6,8-tetramethoxy naphthalene |  | C14H16O4 | 249.1127 | 249.1129 | 5 | 5.63 |
| Gene | Gene Size (nt) | Putative Function | Best BLAST Hit of the Gene Product | ||
|---|---|---|---|---|---|
| Genebank Accession No. | Species | Id/Sim | |||
| orf1 | 780 | voltage-gated potassium channel | PXX59685 | Nocardia tenerifensis | 68/77 |
| orf2 | 729 | hypothetical protein | WP_084160940 | Nocardia sp. sp. BMG51109 | 61/71 |
| thnM1 | 1164 | hypothetical protein | WP_093574921 | Amycolatopsis rubida | 53/66 |
| class I SAM-dependent methyltransferase | WP_018807243 | Salinispora arenicola | 52/65 | ||
| thnT1 | 1293 | MFS transporter | WP_107657235 | Nocardia suismassiliense | 99/99 |
| thnG | 1173 | DUF1205 domain-containing protein | WP_033430793 | Saccharothrix syringae | 53/67 |
| glycosyltransferase | AIW62993 | uncultured bacterium BAC-AB1442/1414/561 | 40/58 | ||
| thnA | 1125 | type III polyketide synthase | WP_045697370 | Streptomyces rubellomurinus | 68/78 |
| thnM2 | 807 | methyltransferase domain-containing protein | WP_084716343 | Saccharothrix syringae | 53/67 |
| thnM3 | 1044 | methyltransferase | WP_033430882 | Saccharothrix syringae | 65/75 |
| Dimerisation domain-containing protein | SDZ58908 | Saccharopolyspora shandongensis | 58/72 | ||
| thnT2 | 1305 | MFS transporter | WP_067522717 | Nocardia uniformis | 87/91 |
| orf3 | 1371 | MBL fold metallo-hydrolase | WP_067522716 | Nocardia uniformis | 80/88 |
| Acetyl-CoA | Malonyl-CoA | ||
|---|---|---|---|
| Strains | Day | (µmol/g DCW) | (µmol/g DCW) |
| Nocardia sp. CS682 | 3 | 9.24 ± 0.48 | 3.46 ± 0.12 |
| 6 | 22.16 ± 5.54 | 38.84 ± 1.60 | |
| Nocardia sp. CS682DR | 3 | 24. 64 ± 1.02 | 18.40 ± 0.96 |
| 6 | 140.32 ± 2.46 | 174.78 ± 3.68 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, R.; Dhakal, D.; Han, J.M.; Lim, H.N.; Jung, H.J.; Yamaguchi, T.; Sohng, J.K. Production of a Novel Tetrahydroxynaphthalene (THN) Derivative from Nocardia sp. CS682 by Metabolic Engineering and Its Bioactivities. Molecules 2019, 24, 244. https://doi.org/10.3390/molecules24020244
Mishra R, Dhakal D, Han JM, Lim HN, Jung HJ, Yamaguchi T, Sohng JK. Production of a Novel Tetrahydroxynaphthalene (THN) Derivative from Nocardia sp. CS682 by Metabolic Engineering and Its Bioactivities. Molecules. 2019; 24(2):244. https://doi.org/10.3390/molecules24020244
Chicago/Turabian StyleMishra, Ravindra, Dipesh Dhakal, Jang Mi Han, Haet Nim Lim, Hye Jin Jung, Tokutaro Yamaguchi, and Jae Kyung Sohng. 2019. "Production of a Novel Tetrahydroxynaphthalene (THN) Derivative from Nocardia sp. CS682 by Metabolic Engineering and Its Bioactivities" Molecules 24, no. 2: 244. https://doi.org/10.3390/molecules24020244