1. Introduction
Gentiana rigescens Franchet (Dian long dan) is a herbaceous species that grows in mountainous regions of Yunnan-Guizhou Plateau in the southwest of China [
1]. Like European traditional medicinal plant yellow gentian (
G. lutea L),
G. rigescens is famous for its bitter properties that are due to the bitter active principles (e.g., loganin, gentiopicroside, swertiamarin, sweroside, etc.) [
2,
3,
4]. Those compounds have pharmacological effects of anti-inflammation, antioxidant, anti-cancer, antiviral, cholagogic agent, hepatoprotective, wound-healing activities, and so forth [
3,
5]. Additionally, they are used to stimulate appetite and improve digestion [
5,
6,
7]. In addition, a series of neuritogenic compounds had been isolated from the aerial and underground parts of
G. rigescens, which could be used as raw material for the preparation of functional food and a therapeutic drug for Alzheimer’s disease [
8,
9,
10,
11]. Now,
G. rigescens have been the official drug of Chinese pharmacopoeia (2015 edition) for chronic hepatitis and important raw materials for the pharmaceutical industry in China [
12].
G. rigescens were usually collected from different regions of Yunnan-Guizhou Plateau in order to provide satisfaction of continuously increasing industrial demands for raw materials. However, some of the researchers had reported that chemical constitutions of underground part of
G. rigescens were extremely variable and diverse according to plant grown location or producing area [
13,
14,
15]. Quantitative analysis of bioactivity compounds (such as gentiopicroside, sweroside, swertiamarin, isoorientin, and other compounds) from rhizomes, stems, leaves, and flowers indicated that northwest of Yunnan-Guizhou Plateau was suitable for chemical compounds accumulation [
13,
14,
15,
16]. Additionally, conversion and transport of those compounds might be influenced by climatic conditions in the plant habitat [
14,
17].
Latitude has a strong impact on the local climate environment in southwest China [
18,
19]. As the main distribution area of
G. rigescens, Yunnan-Guizhou Plateau is characterized by very complex topography and it displays a wide variety of micro-climates [
18,
19,
20,
21]. There are six climatic zones from the north towards the south [
20]. Especially, in the higher latitude areas, such as northwest Yunnan or south of the Hengduan Mountains (26–28° N), the temperature gradients are more abrupt than in the other regions [
19]. Furthermore, precipitation and temperature in the Yunnan-Guizhou Plateau also show clear variations along the latitude gradients [
19,
21]. Therefore, it is necessary to explore the variation of phytochemical and medicinal material quality of
G. rigescens that were grown in different latitudes and build a classification model for tracing producing areas of medicinal materials.
As we know, the contents of bioactive compounds and quality of medicinal materials have a close relationship with the environment of producing area [
22,
23,
24,
25]. Quality control and geographical indication of medicinal materials raise many concerns by pharmaceutical industries with the expansion in the use of herbal medicines. However, using few marker compounds could not reflect the chemical complexity of herbs and this method is hard to effectively authenticate the origin of herbal medicines [
26,
27]. Chemical fingerprints, as a comprehensive evaluation methodology, have been widely used to deal with the problem [
26,
28,
29]. In recent years, infrared spectroscopy (IR), UV-Vis spectroscopy (UV-Vis), and other spectral fingerprints have been well-established analytical techniques for geographical traceability studies of
G. rigescens and other medicinal plants in the worldwide [
30,
31,
32,
33,
34]. In contrast, there were limited reports on the use of chromatographic fingerprint to identify the producing regions of herbal materials [
30,
31,
32,
33,
34,
35]. Although there were many reports about discrimination of herbs according to their producing areas while using liquid chromatography technology, most of them are based on the information of limited chemical markers or chromatographic profiles [
36,
37,
38,
39]. The potential of chromatographic fingerprints for herbs authentication needs to be further explored.
When compared with chemical marker or chromatographic profile (targeted), chromatographic fingerprint (untargeted) contains unspecific and non-evident information and chemometric tools should extract chemical information [
40]. Recently, literature reported some successful studies applying chromatographic fingerprint, together with chemometric methodology, to discriminate herbs and food samples of different origin or cultivars [
41,
42,
43,
44]. All of those studies suggested that it is possible to develop a reliable and accurate method for the geographical tracing of
G. rigescens by applying the chromatographic fingerprint methodology.
In the progression of improving geographical authentication of food and drugs, one of the important goals is building discrimination models with a less error rate and reducing the uncertainty of the prediction results [
33,
44]. Data fusion strategy has been widely used in the last years in the field of food authentication in order to improve class discrimination techniques [
45]. Some reports about
Panax notoginseng,
Paris Polyphylla var.
yunnanensis and other herb materials also showed the huge potential of this strategy in the discrimination of medicinal materials producing areas [
46,
47,
48]. Today, most of the fused data come from spectral fingerprint and very few studies report the data fusion of chromatographic fingerprint [
42,
43]. Furthermore, data fusion studies are mostly based on the fusion of multivariate instrumental techniques [
42,
43], while reports of
P. Polyphylla var.
yunnanensis,
Macrohyporia cocos, and other species indicated that reliable classification results were also available by the fusion analysis of chemical fingerprint data collected from different medicinal parts of herbs [
35,
49]. Accumulation and distribution of metabolites in the different parts of plants were different because of the differential response of root, stem, flower and other organs to the environment variation of producing area [
17,
50]. Therefore, fingerprint data fusion of multi-medicinal parts may provide integrated chemical information for the authentication of medicinal materials. At the same time, this method also contributes to a more comprehensive understanding of the response and adaptation of medicinal plants to complex geographical environments.
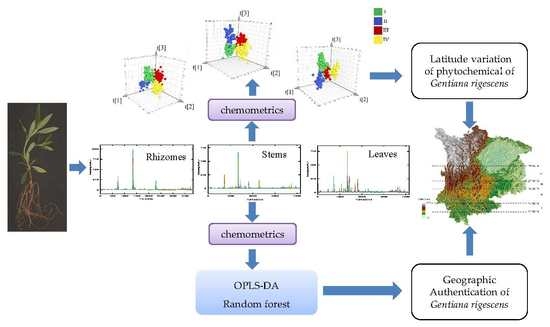
The aim of this study is to explore the variation of chromatographic fingerprints of G. rigescens along the latitude gradients and to use chemometrics to mine fingerprint chemical information, and to investigate the potential of the untargeted chromatographic fingerprint to trace herbs grown at different latitudes. For this purpose, we developed fingerprint of rhizomes, stems, and leaves of G. rigescens by high-performance liquid chromatography with diode array detection (HPLC-DAD) technology. Subsequently, classification models for the identification of different producing areas were built by HPLC fingerprint combined with RF (random forest algorithm) and OPLS-DA (orthogonal partial least-squares discriminant analysis). At last, two types of data fusion strategies, “low- level” and “mid-level” data fusion, were studied in order to improve the model performances.
3. Materials and Methods
3.1. Plant Material Collection
Plant materials (29 population and 280 individuals) of
G. rigescens were collected in the fall of 2012 and 2013 at the time of local traditional harvest period, at the different location of Yunnan, Guizhou, and Sichuan (
Figure 12). Four producing areas were divided according to the location of population. (I) low latitudes area, with latitudes ranging from 23.92–23.66° N, South of Yunnan (eight population and 76 individuals), (II) mid-latitude area, with latitudes ranges from 24.95–25.06° N, Middle of Yunnan (five population and 48 individuals), (III) mid-high latitude area, with latitudes ranges from 26.49–26.64° N, Northwest of Yunnan and West of Guizhou (nine population and 76 individuals 87), and (IV) high latitude area, with latitudes ranges from 27.34–28.52° N, Hengduan Mountains Region of Yunnan and mountainous regions of Southwest of Sichuan (seven population and 69 individuals). The fresh materials were authenticated and transported to the laboratory of Yuxi normal University. Subsequently, samples were wash cleaning and dried at 50 °C as soon as possible. At last, all samples (rhizomes, stems and leaves) were stored in a relatively dry environment prior to the extraction procedure.
3.2. Chemicals and Reagents
HPLC-grade acetonitrile, methanol (MeOH) were supplied by Thermo Fisher Scientific (Waltham, MA, USA). HPLC-grade formic acid was purchased from Sigma-Aldrich (Steinheim, Germany). Deionized water was obtained from Wahaha Group Co., Ltd. (Hangzhou, Zhejiang, China). The primary grade reference standards loganin (purity: ≥98%), 6′-O-β-d-glucopyranosylgentiopicroside (purity: ≥98%), swertiamarine (purity: ≥98%), gentiopicroside (purity: ≥98%), and sweroside (purity: ≥98%) were purchased from the Chinese National Institute for Food and Drug Control (Beijing, China), Shanghai Shifeng Biological Technology (Shanghai, China), respectively.
3.3. Sample Preparation
The dried samples (rhizomes, stems, and leaves) were ground and then passed through a 100 mesh sieves. Each sample powder (25 mg) was accurately weighed and extracted while using 1.5 mL 80% methanol-water solution, at 25 °C. The samples were extracted while using an Ultrasonic extractor for 40 min. The final extract was filtered with a 0.22 μm syringe filter into an HPLC vial and then subjected to HPLC analysis [
16,
58].
3.4. Instrumentation and HPLC Analysis
Chromatographic analyses were performed with an Agilent 1260 Infinity LC system (Agilent Technologies, Santa Clara, CA, USA), which was equipped with a G1315D diode-array detector, a G1329B ALS autosampler, and a thermostated column compartment. The HPLC fingerprint was recorded by Chemstation software (Agilent Technologies, Waldbron, Germany).
The analytical separation was adopted from a published method for chemical fingerprinting analysis [
16]. The separation was achieved on a reversed phase C18 (Agilent Intersil, 5 µm, 4.6 × 150 mm) column (Agilent, Santa Clara, CA, USA). The composition of the mobile phase was: (A) 0.1% phosphoric acid in water and (B) 100% acetonitrile. The separation was as follows: 0.00–2.50 min: 7–10% B, 2.50–20.00 min: 10–26% B, 20.00–29.02 min: 26–58.3% B, 29.02–30.00 min: 58.3–90% B. The column was subsequently washed with 90% B and re-equilibrated with 7% B prior to injection of the next sample. The flow rate was 1.0 mL/min and the column temperature was 30 °C. The injection volume was 5 µL and the detective wavelength of UV spectra was set at 241 nm. Chromatographic data was processed while using OpenLab software (Agilent Technologies) [
16,
58].
3.5. Data Analysis
HPLC fingerprints from the 280 rhizome samples, 280 stem samples, and 280 leaf samples, a total of 840 fingerprint data was exported in CSV format and imported to MATLAB R2018b (The MathWorks, Inc., Natick, MA, USA), which was used for correlation optimized warping (COW) alignment preprocessing of chromatographic fingerprint. MATLAB code of COW is freely available from
www.models.kvl.dk. The preprocessing fingerprint was analyzed in the following work [
59].
Exploratory data analysis (EDA) is necessary for building predictive models [
60,
61]. It can help in determining interesting correlations among all of the samples or variables and summarize data sets main characteristics [
60]. Principal component analysis (PCA) is a popular primary tool in EDA [
61,
62]. It is often used to visualize the relatedness between samples and explains the variance in the data. Hence, PCA, as an unsupervised pattern recognition technique, was widely used to extract key information from chemical fingerprint for geographical origin or Modelling Research [
61].
Unlike PCA, orthogonal partial least squares discriminant analysis (OPLS-DA) is a supervised pattern recognition technique. As an extension of PLS, an inbuilt orthogonal signal correction filter was incorporated in the OPLS-DA model [
56]. This algorithm effectively divides the X variable into two parts: one part that is related to class information (Y-predictive) and the other is orthogonal or unrelated to class information (Y-uncorrelated). Therefore, interpretability and prediction performance of the model was enhanced [
56].
Random forest (RF) is another supervised pattern recognition technique utilized in the study. RF is an ensemble learning method [
55]. A large number of trees were produced by RF algorithm in order to improve model predictive ability, and trees’ decision results were combined as final decision results. In other words, the more trees built in the random forest classifier, the higher accuracy could be achieved. However, many researches showed that an optimum tree number was of great importance in modeling classification performance [
33,
46].
In this work, exploratory data analysis of HPLC fingerprints of
G. rigescens grown in four different latitudes was finished with PCA. Two supervised pattern recognition techniques, OPLS-DA and RF, were applied to build classification models for
G. rigescens producing areas. SIMCA 14.1 software managed PCA and OPLS-DA (Umetrics AB, Umea, Sweden). RF classification models were established with R 3.5.1 program and package randomForest (Version 4.6-14) [
63].
Data Fusion Strategy
In the case of low-level fusion strategy (
Figure 11), different subsets HPLC fingerprint data matrix of rhizomes, stems, and leaves) are straightforwardly concatenated and compiled into a new chromatographic data matrix for subsequent classification model construction [
45,
46]. Furthermore, each subset must be totally aligned and keep all the variables on the same scale before subsets reconnection [
45,
46].
In the case of mid-level fusion (
Figure 11), the first step of data treatment is feature selection that is based on rhizomes, stems, or leaves classification models. When compared with the raw data sets, feature selection of subsets minimizes the data content and reduces data dimensions. Subsequently, new subsets of rhizomes, stems, and leaves were rebuilt while using variables of feature selection [
45]. At last, those subsets are concatenated and compiled into a final data matrix for model construction [
45].
In the research, relevant variables of RF classification models were determined by the R software package Boruta [
64], and VIP was used for important variables selection of OPLS-DA [
65].
3.6. Model Evaluation
Five parameters, including accuracy (ACC), sensitivity (SE), specificity (SP), efficiency (EFF), and Matthews correlation coefficient (MCC) were applied to evaluate the identification ability of RF and OPLS-DA models. The ruggedness of OPLS-DA model was investigated through 200 times permutation tests. Furthermore, cumulative prediction ability (Q
2), cumulative interpretation ability (R
2), root mean square error of estimation (RMSEE), root mean square error of cross-validation (RMSECV), and root mean square error of prediction (RMSEP) were important evaluation indexes for the predictive power of OPLS-DA model [
33,
66].
Values of TP (Correctly identified samples of positive class), TN (correctly identified samples of negative class), FN (incorrectly identified samples of positive class), and FP (incorrectly identified samples of negative class) were calculated according to confusion matrixes of classification models. Subsequently, ACC, SE, SP, EFF, and MCC were calculated while using Equations (1)–(5) and values of Q2, R2, RMSEE, RMSECV, and RMSEP computed by software SIMCA 14.1.
For model performance, lower values of RMSEE, RMSECV, and RMSEP mean better predictive ability for the models. Conversely, the closer that values of ACC, SE, SP, EFF, MCC, and Q2, R2 are to 1, the more well performance the model is.