A Simple Mannose-Coated Poly (p-Phenylene Ethynylene) for Qualitative Bacterial Capturing
Abstract
:1. Introduction
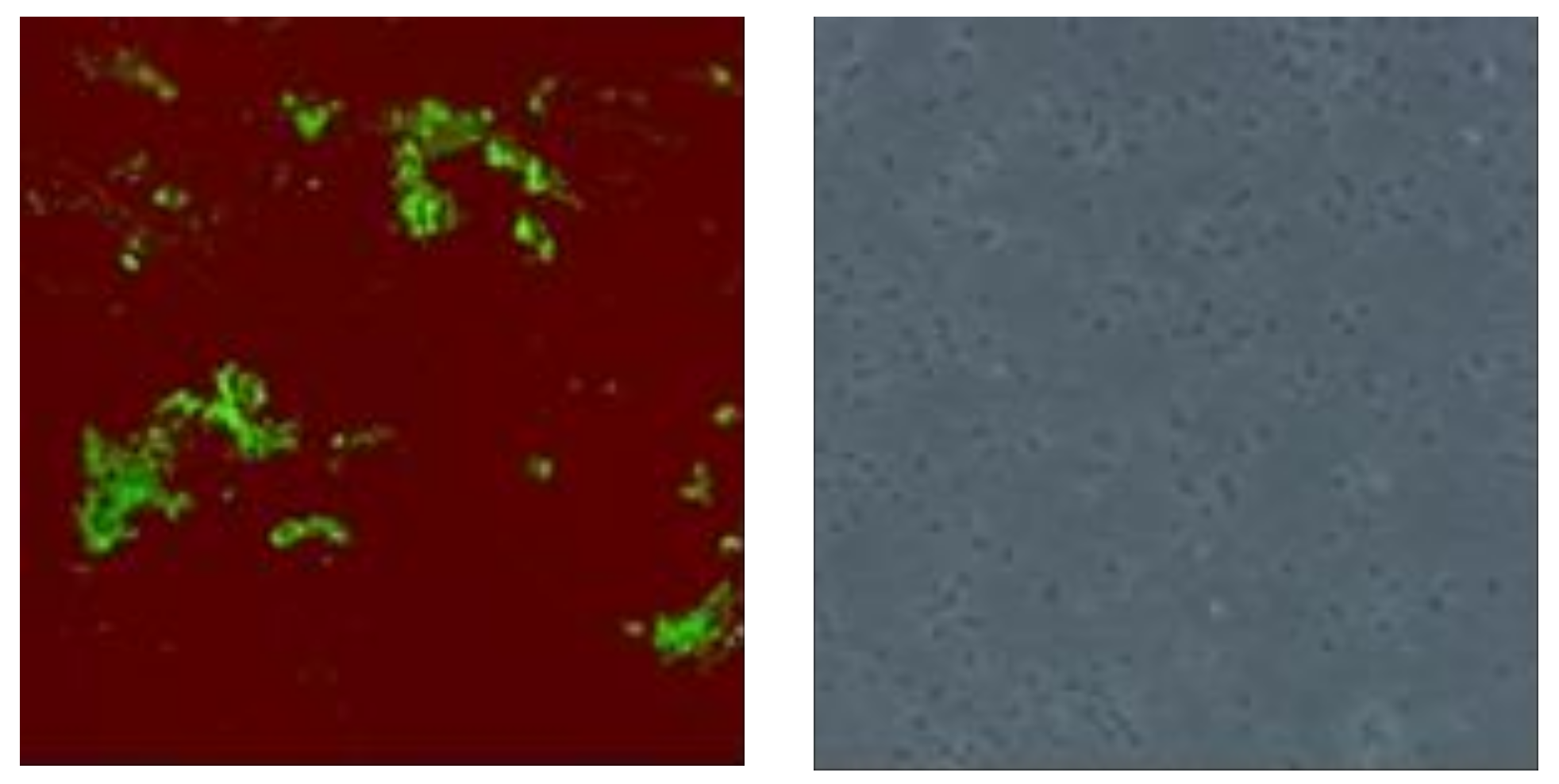
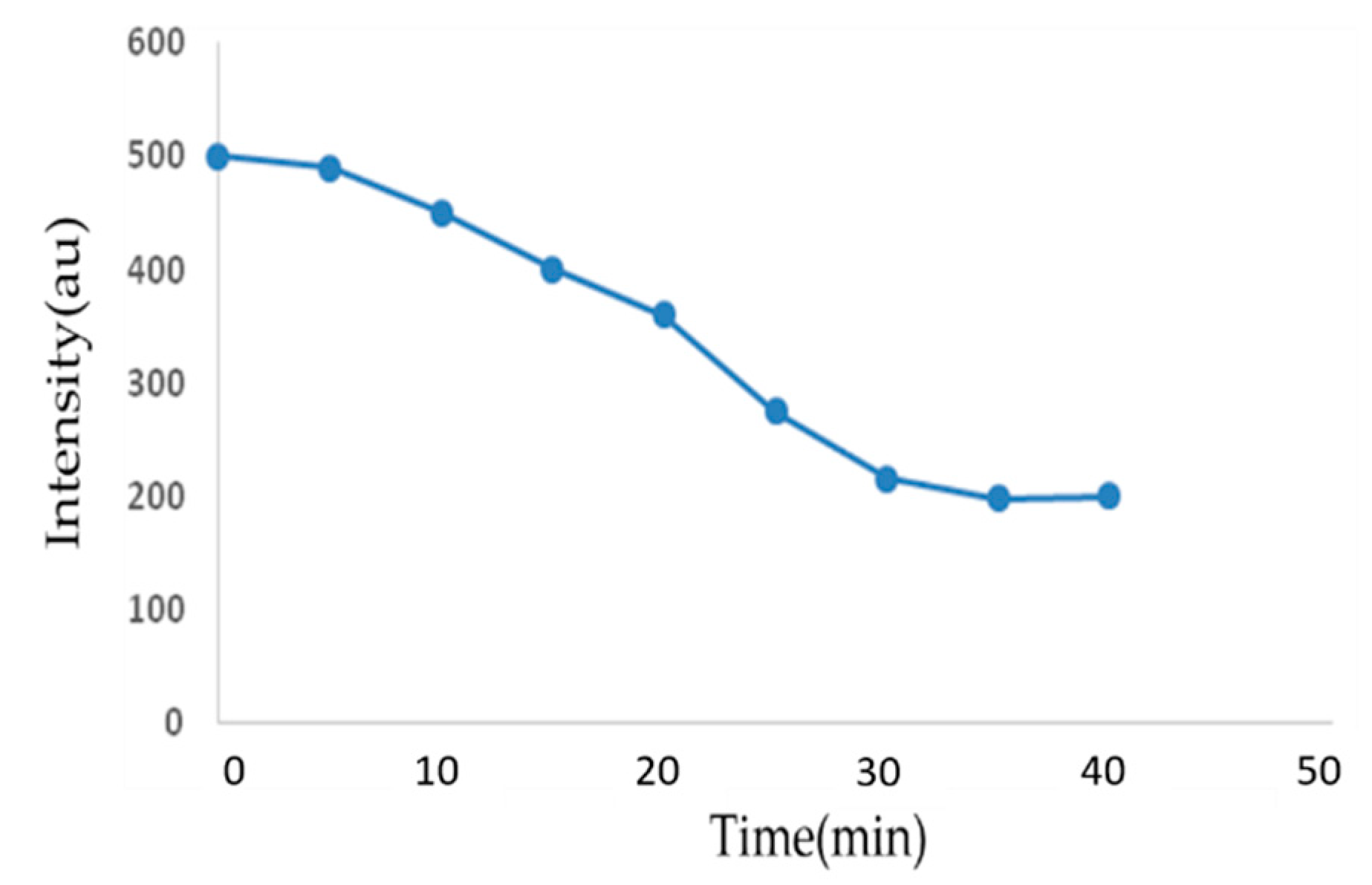
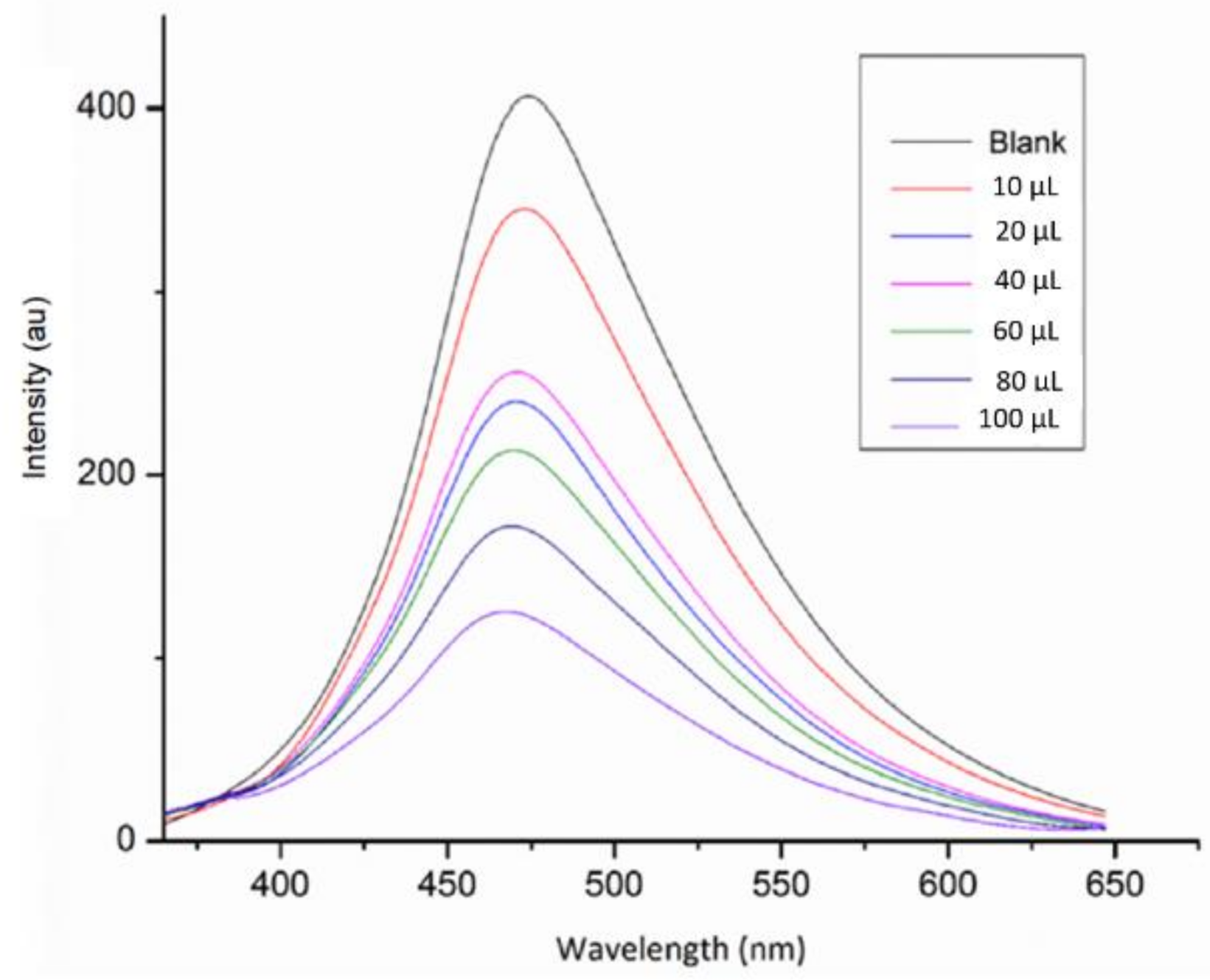
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Polymer Synthesis and Characterization
3.3. Sugar Loading Test
3.4. Cell Culture and Incubation
3.5. Optimum Binding Time
3.6. Sensitivity Test
3.7. Cell Imaging
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carlson, K.; Misra, M.; Mohanty, S. Developments in Micro- and Nanotechnology for Foodborne Pathogen Detection. Foodborne Pathog. Dis. 2018, 15, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, X.; Han, L.; Niu, S.; Shi, C.; Ma, C. Rapid detection of foodborne pathogen Listeria monocytogenes by strand exchange amplification. Anal. Biochem. 2018, 545, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Morschett, H.; Schiprowski, D.; Rohde, J.; Wiechert, W.; Oldiges, M. Comparative evaluation of phototrophic microtiter plate cultivation against laboratory-scale photobioreactors. Bioprocess Biosyst. Eng. 2017, 40, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Jasson, V.; Jacxsens, L.; Luning, P.; Rajkovic, A.; Uyttendaele, M. Alternative microbial methods: An overview and selection criteria. Food Microbiol. 2010, 27, 710–730. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Poshtiban, S.; Evoy, S. Recent Advances in Bacteriophage Based Biosensors for Food-Borne Pathogen Detection. Sensors 2013, 13, 1763–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Sindhu, A.; Kaur, H.; Dilbaghi, N.; Chaudhury, A. An overview of transducers as platform for the rapid detection of foodborne pathogens. Appl. Microbiol. Biotechnol. 2013, 97, 1829–1840. [Google Scholar] [CrossRef] [PubMed]
- Xu, S. Electromechanical biosensors for pathogen detection. Microchimica Acta 2012, 178, 245–260. [Google Scholar] [CrossRef]
- Amiri, M.; Bezaatpour, A.; Jafari, H.; Boukherroub, R.; Szunerits, S. Electrochemical Methodologies for the Detection of Pathogens. ACS Sens. 2018, 3, 1069–1086. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Aydin, M.; Khatiwara, A.; Dolan, M.C.; Gilmore, D.F.; Bouldin, J.L.; Ahn, S.; Ricke, S.C. Current and emerging technologies for rapid detection and characterization of Salmonella in poultry and poultry products. Food Microbiol. 2014, 38, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Seckute, J.; Devaraj, N.K. Expanding room for tetrazine ligations in the in vivo chemistry toolbox. Curr. Opin. Chem. Biol. 2013, 17, 761–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharon, N. Bacterial Lectins, Cell-Cell Recognition and Infectious-Disease. FEBS Lett. 1987, 217, 145–157. [Google Scholar] [CrossRef]
- Lis, H.; Sharon, N. Lectins: Carbohydrate-specific proteins that mediate cellular recognition. Chem. Rev. 1998, 98, 637–674. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Sasada, M.; Yamanaka, M.; Nasu, M. Rapid detection of respiring Escherichia coli O157:H7 in apple juice, milk, and ground beef by flow cytometry. Cytom. Part A J. Int. Soc. Anal. Cytol. 2003, 54, 27–35. [Google Scholar]
- Yu, L.S.L.; Reed, S.A.; Golden, M.H. Time-resolved fluorescence immunoassay (TRFIA) for the detection of Escherichia coli O157:H7 in apple cider. J. Microbiol. Methods 2002, 49, 63–68. [Google Scholar] [CrossRef]
- Goodridge, L.; Chen, J.R.; Griffiths, M. The use of a fluorescent bacteriophage assay for detection of Escherichia coli O157:H7 in inoculated ground beef and raw milk. Int. J. Food Microbiol. 1999, 47, 43–50. [Google Scholar] [CrossRef]
- Disney, M.D.; Seeberger, P.H. The use of carbohydrate microarrays to study carbohydrate-cell interactions and to detect pathogens. Chem. Biol. 2004, 11, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, B.; Martins, M.B.; Karamanska, R.; Russell, D.A.; Field, R.A. Bacterial detection using carbohydrate-functionalised CdS quantum dots: A model study exploiting E. coli recognition of mannosides. Tetrahedron Lett. 2009, 50, 886–889. [Google Scholar] [CrossRef]
- Huang, A.H.; Qiu, Z.G.; Jin, M.; Shen, Z.Q.; Chen, Z.L.; Wang, X.W.; Li, J.W. High-throughput detection of food-borne pathogenic bacteria using oligonucleotide microarray with quantum dots as fluorescent labels. Int. J. Food Microbiol. 2014, 185, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Torun, O.; Boyaci, I.H.; Temur, E.; Tamer, U. Comparison of sensing strategies in SPR biosensor for rapid and sensitive enumeration of bacteria. Biosens. Bioelectron. 2012, 37, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Lazaro, D.; D’Agostino, M.; Herrewegh, A.; Pla, M.; Cook, N.; Ikonomopoulos, J. Real-time PCR-based methods for detection of Mycobacterium avium Subsp paratuberculosis in water and milk. Int. J. Food Microbiol. 2005, 101, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Green, H.C.; Field, K.G. Sensitive detection of sample interference in environmental qPCR. Water Res. 2012, 46, 3251–3260. [Google Scholar] [CrossRef] [PubMed]
- Lazcka, O.; Del Campo, F.J.; Munoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007, 22, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Pan, C.Y.; Luo, M.D.; Zhang, H.B. Fluorescent Mannose-Functionalized Hyperbranched Poly (amido amine)s: Synthesis and Interaction with E. coli. Biomacromolecules 2010, 11, 1840–1846. [Google Scholar] [CrossRef] [PubMed]
- Arias, E.; Mendez, M.T.; Arias, E.; Moggio, I.; Ledezma, A.; Romero, J.; Margheri, G.; Giorgetti, E. Supramolecular Recognition of Escherichia coli Bacteria by Fluorescent Oligo(Phenyleneethynylene)s with Mannopyranoside Termini Groups. Sensors 2017, 17, 1025. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.L.; Kim, I.B.; Tolbert, L.M.; Bunz, U.H.F. Fluorescence self-quenching of a mannosylated poly(p-phenyleneethynylene) induced by Concanavalin A. J. Am. Chem. Soc. 2008, 130, 6952–6954. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.B.; Meng, J.; Dong, Y.; Cheng, Y.X.; Zhu, C.J. Polymer-based fluorescence sensor incorporating triazole moieties for Hg2+ detection via click reaction. Polymer 2010, 51, 3064–3067. [Google Scholar] [CrossRef]
- Thomas, S.W.; Joly, G.D.; Swager, T.M. Chemical sensors based on amplifying fluorescent conjugated polymers. Chem. Rev. 2007, 107, 1339–1386. [Google Scholar] [CrossRef] [PubMed]
- Wosnick, J.H.; Mello, C.M.; Swager, T.M. Synthesis and application of poly (phenylene ethynylene)s for bioconjugation: A conjugated polymer-based fluorogenic probe for proteases. J. Am. Chem. Soc. 2005, 127, 3400–3405. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Q.; Fan, Q.L.; Zhang, G.W.; Chen, Y.; Lu, X.M.; Huang, W. A fluorene-containing water-soluble poly(p-phenyleneethynylene) derivative: Highly fluorescent and sensitive conjugated polymer with minor aggregation in aqueous solution. Polymer 2006, 47, 5233–5238. [Google Scholar] [CrossRef]
- Pinto, M.R.; Kristal, B.M.; Schanze, K.S. A water-soluble poly (phenylene ethynylene) with pendant phosphonate groups. Synthesis, photophysics, and layer-by-layer self-assembled films. Langmuir 2003, 19, 6523–6533. [Google Scholar] [CrossRef]
- Yang, S.K.; Shi, X.H.; Park, S.; Ha, T.; Zimmerman, S.C. A dendritic single-molecule fluorescent probe that is monovalent, photostable and minimally blinking. Nat. Chem. 2013, 5, 692–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, C.P.; Dierckx, A.; Miannay, F.A.; Wellner, E.; Wilhelmsson, L.M.; Grotli, M. Synthesis and photophysical characterisation of new fluorescent triazole adenine analogues. Org. Biomol. Chem. 2014, 12, 5158–5167. [Google Scholar] [CrossRef] [PubMed]
- Kenmoku, S.; Urano, Y.; Kojima, H.; Nagano, T. Development of a highly specific rhodamine-based fluorescence probe for hypochlorous acid and its application to real-time imaging of phagocytosis. J. Am. Chem. Soc. 2007, 129, 7313–7318. [Google Scholar] [CrossRef] [PubMed]
- Tsakama, M.; Shang, Y.T.; He, Y.H.; Fan, B.; Wang, F.Z.; Chen, W.H.; Dai, X.F. Synthesis and optical properties of a novel sugar coated poly(p-phenyleneethynylene) effectively quenched by concanavalin A. Tetrahedron Lett. 2016, 57, 1739–1742. [Google Scholar] [CrossRef]
- Thavornsin, N.; Sukwattanasinitt, M.; Wacharasindhu, S. Direct synthesis of poly (p-phenyleneethynylene)s from calcium carbide. Polym. Chem. 2014, 5, 48–52. [Google Scholar] [CrossRef]
- Lee, K.; Kim, H.J.; Kim, J. Design Principle of Conjugated Polyelectrolytes to Make Them Water-Soluble and Highly Emissive. Adv. Funct. Mater. 2012, 22, 1076–1086. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.Y.; Pinto, M.R.; Schanze, K.S. Photophysics, aggregation and amplified quenching of a water-soluble poly (phenylene ethynylene). Chem. Commun. 2002, 7, 446–447. [Google Scholar] [CrossRef]
- Zeng, X.Q.; Andrade, C.A.S.; Oliveira, M.D.L.; Sun, X.L. Carbohydrate-protein interactions and their biosensing applications. Anal. Bioanal. Chem. 2012, 402, 3161–3176. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |



| MW/gmol−1 | Sugar Loading (%) | λ abs/nm | λ em/nm | Stokes Shift (cm−1) | ΦF (%) |
|---|---|---|---|---|---|
| >14,000 | 30 | 393 | 466.5 | 73.3 | 10.8 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsakama, M.; Ma, X.; He, Y.; Chen, W.; Dai, X. A Simple Mannose-Coated Poly (p-Phenylene Ethynylene) for Qualitative Bacterial Capturing. Molecules 2018, 23, 2056. https://doi.org/10.3390/molecules23082056
Tsakama M, Ma X, He Y, Chen W, Dai X. A Simple Mannose-Coated Poly (p-Phenylene Ethynylene) for Qualitative Bacterial Capturing. Molecules. 2018; 23(8):2056. https://doi.org/10.3390/molecules23082056
Chicago/Turabian StyleTsakama, Madalitso, Xiaochi Ma, Yonghuan He, Weihua Chen, and Xiaofeng Dai. 2018. "A Simple Mannose-Coated Poly (p-Phenylene Ethynylene) for Qualitative Bacterial Capturing" Molecules 23, no. 8: 2056. https://doi.org/10.3390/molecules23082056




