A New Method for Recognizing Cytokines Based on Feature Combination and a Support Vector Machine Classifier
Abstract
:1. Introduction
2. Classifier and Verification Methods
3. Measurements
4. Feature Combinations and Results
4.1. Performance of Feature Methods
4.2. Feature Combinations
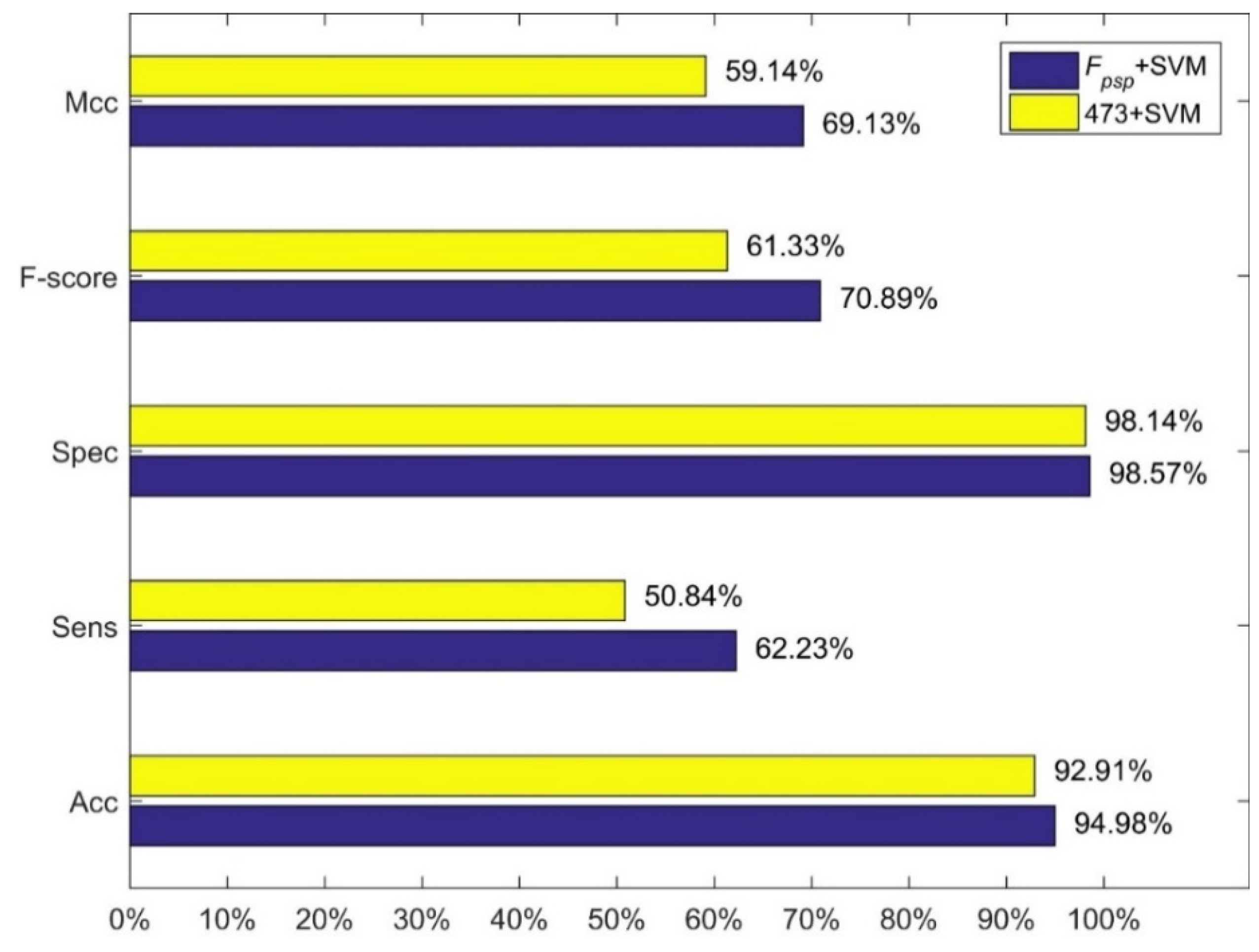
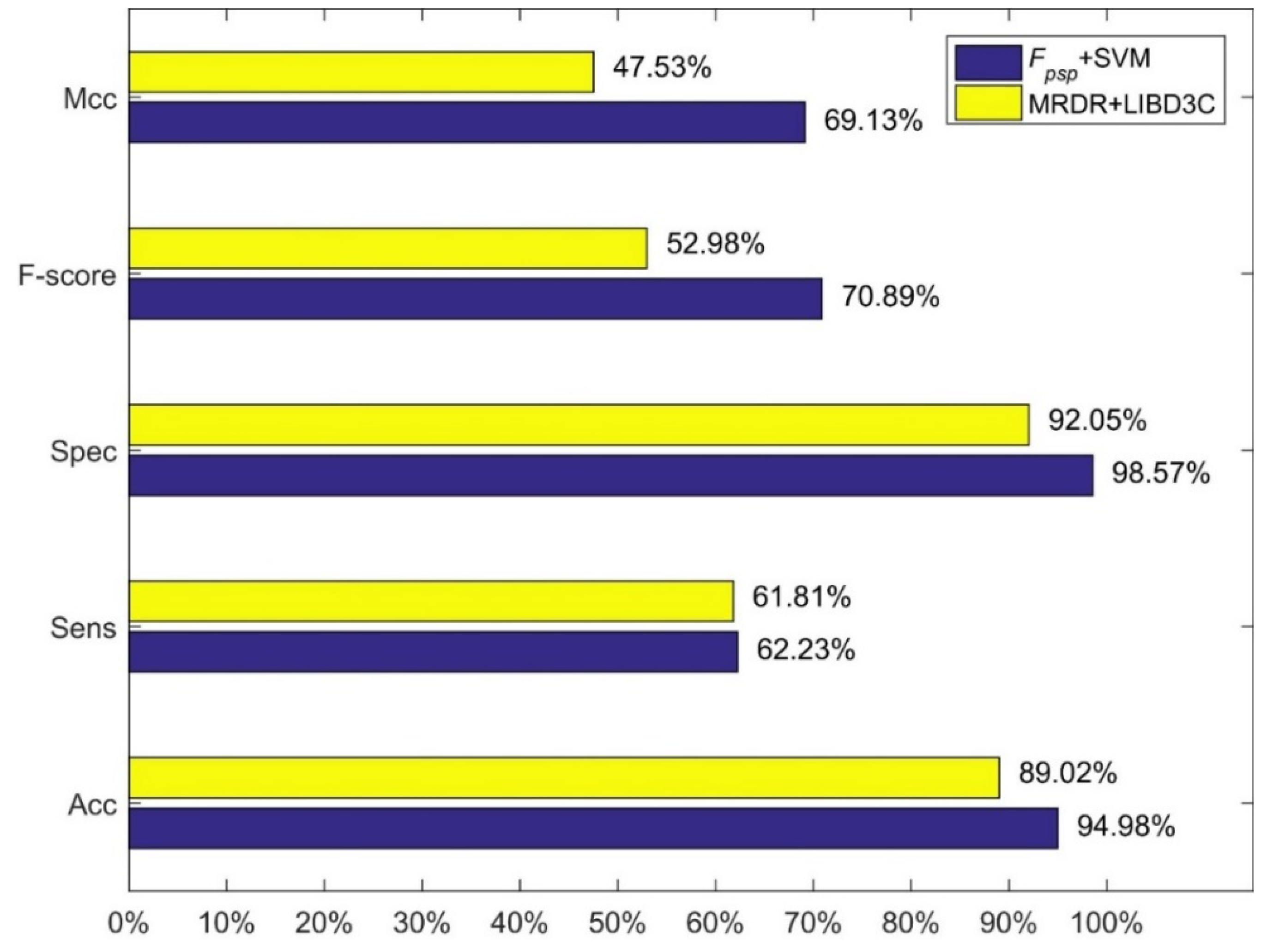
4.3. Comparison with Other Methods
5. Discussion
6. Conclusions
7. Materials and Methods
7.1. Data
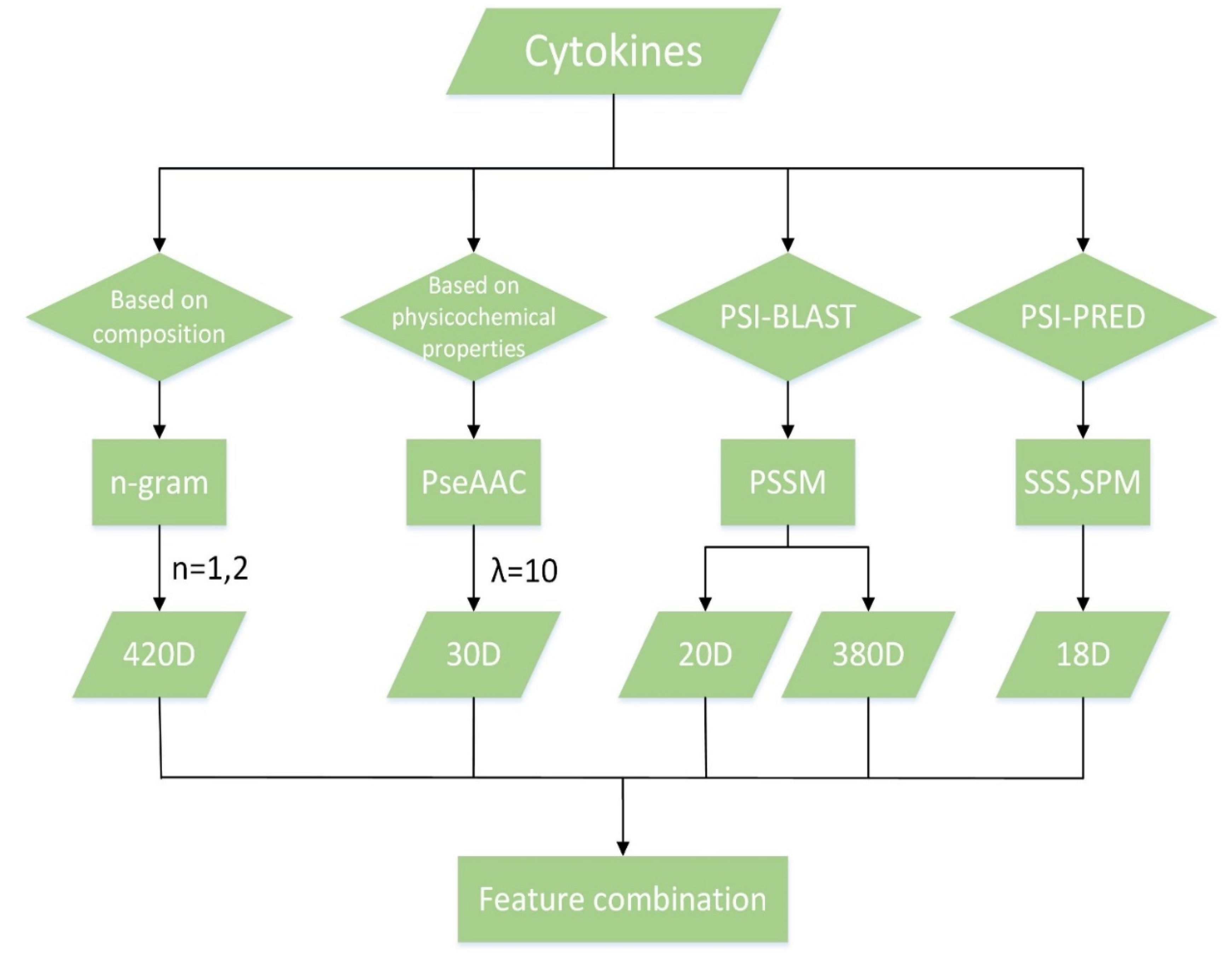
7.2. Feature Extraction
7.2.1. n-Gram
7.2.2. PseAAC
7.2.3. Secondary Structure
Author Contributions
Funding
Conflicts of Interest
References
- Sutovsky, J.; Kocmalova, M.; Benco, M.; Kazimierova, I.; Pappova, L.; Frano, A.; Sutovska, M. The role of cytokines in degenerative spine disorders. Eur. Pharm. J. 2017, 64, 26–29. [Google Scholar] [CrossRef] [Green Version]
- Vandergeeten, C.; Fromentin, R.; Chomont, N. The role of cytokines in the establishment, persistence and eradication of the hiv reservoir. Cytokine Growth Factor Rev. 2012, 23, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Si, M.; Jiao, X.; Li, Y.; Chen, H.; He, P.; Jiang, F. The role of cytokines and chemokines in the microenvironment of the blood–brain barrier in leukemia central nervous system metastasis. Cancer Manag. Res. 2018, 10, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Musolino, C.; Allegra, A.; Innao, V.; Allegra, A.G.; Pioggia, G.; Gangemi, S. Inflammatory and anti-inflammatory equilibrium, proliferative and antiproliferative balance: The role of cytokines in multiple myeloma. Mediat. Inflamm. 2017, 2017, 1852517. [Google Scholar] [CrossRef] [PubMed]
- Champsi, J.H.; Bermudez, L.E.; Young, L.S. The role of cytokines in mycobacterial infection. Biotherapy 1994, 7, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Gnanadurai, C.W.; Zhen Fang, F.U. Critical roles of chemokines and cytokines in antiviral innate immune responses during rabies virus infection. Front. Agric. Sci. Eng. 2017, 4, 260–267. [Google Scholar] [CrossRef]
- Nakajima, H.; Takatsu, K. Role of cytokines in allergic airway inflammation. Int. Arch. Allergy Immunol. 2007, 142, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.J.; Tao, L.; Lu, L.M.; Tang, D.; Shi, X.L. Role of t helper 17 cytokines in the tumour immune inflammation response of patients with laryngeal squamous cell carcinoma. Oncol. Lett. 2017, 14, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, H.; Nishikawa, K. Discrimination of intracellular and extracellular proteins using amino acid composition and residue-pair frequencies. J. Mol. Boil. 1994, 238, 54. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.Y.; Feng, Z.P.; Liu, J.K. Prediction of protein structural class by amino acid and polypeptide composition. Eur. J. Biochem. 2010, 269, 4219–4225. [Google Scholar] [CrossRef]
- Shen, H.; Chou, K. Ensemble classifier for protein fold pattern recognition. Bioinformatics 2006, 22, 1717–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–4002. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, L.; Lv, J. Accurate prediction of protein structural classes by incorporating predicted secondary structure information into the general form of chou’s pseudo amino acid composition. J. Theor. Boil. 2014, 344, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Kong, L.; Han, X.; Lv, J. Structural class prediction of protein using novel feature extraction method from chaos game representation of predicted secondary structure. J. Theor. Boil. 2016, 400, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices 1. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Kamal, N.A.M.; Bakar, A.A.; Zainudin, S. Classification of human membrane protein types using optimal local discriminant bases feature extraction method. J. Theor. Appl. Inf. Technol. 2018, 96, 767–771. [Google Scholar]
- Zhang, S.; Duan, X. Prediction of protein subcellular localization with oversampling approach and chou’s general pseaac. J. Theor. Boil. 2018, 437, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.K.; Namdev, N.; Kumar, A. Rough set method accurately predicts unknown protein class/family of leishmania donovani membrane proteome. Math. Biosci. 2018, 301, 37. [Google Scholar] [CrossRef] [PubMed]
- Huo, H.; Yang, L. Prediction of conotoxin superfamilies by the naive bayes classifier. In Proceedings of the 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Shanghai, China, 14–16 October 2017. [Google Scholar]
- Vapnik, V.N. The nature of statistical learning theory. IEEE Trans. Neural Netw. 1997, 38, 409. [Google Scholar]
- Rahman, J.; Mondal, M.N.; Islam, M.K.; Hasan, M.A. Feature fusion based svm classifier for protein subcellular localization prediction. J. Integr. Bioinform. 2016, 13, 23–33. [Google Scholar] [CrossRef]
- Mei, J.; Ji, Z. Prediction of hiv-1 and hiv-2 proteins by using chou’s pseudo amino acid compositions and different classifiers. Sci. Rep. 2018, 8, 2359. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Tuck, D.P. Msvm-rfe: Extensions of svm-rfe for multiclass gene selection on DNA microarray data. Bioinformatics 2007, 23, 1106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Zhang, H.; Jiang, X.; Xu, B.; Xue, Y.; Cao, Y.; Zhai, Q.; Zhai, Y.; Xu, M. Prediction of novel pre-micrornas with high accuracy through boosting and svm. Bioinformatics 2011, 27, 1436–1437. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Sarai, A. Pssm-based prediction of DNA binding sites in proteins. BMC Bioinform. 2005, 6, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, N.; Chen, H.; Sun, Z. Ctkpred: An svm-based method for the prediction and classification of the cytokine superfamily. Protein Eng. Des. Sel. PEDS 2005, 18, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Yuan, S.; Huang, X.; Zou, Q. Identification of cytokine via an improved genetic algorithm. Front. Comput. Sci. 2015, 9, 643–651. [Google Scholar] [CrossRef]
- Jiang, L.; Liao, Z.; Su, R.; Wei, L. Improved identification of cytokines using feature selection techniques. Lett. Org. Chem. 2017, 14, 632–641. [Google Scholar] [CrossRef]
- Yang, B.; Wu, Q.; Ying, Z.; Sui, H. Predicting protein secondary structure using a mixed-modal svm method in a compound pyramid model. Knowl.-Based Syst. 2011, 24, 304–313. [Google Scholar] [CrossRef]
- Wei, Z.S.; Han, K.; Yang, J.Y.; Shen, H.B.; Yu, D.J. Protein-protein interaction sites prediction by ensembling svm and sample-weighted random forests. Neurocomputing 2016, 193, 201–212. [Google Scholar] [CrossRef]
- Krajewski, Z.; Tkacz, E. Feature selection of protein structural classification using svm classifier. Biocybern. Biomed. Eng. 2013, 33, 47–61. [Google Scholar] [CrossRef]
- Bhasin, M.; Raghava, G.P. Gpcrpred: An svm-based method for prediction of families and subfamilies of g-protein coupled receptors. Nucleic Acids Res. 2004, 32, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Lin, C.J. Libsvm: A Library for Support Vector Machines; ACM: New York, NY, USA, 2011; pp. 1–27. [Google Scholar]
- BW, M. Comparison of the predicted and observed secondary structure of t4 phage lysozyme. BBA—Protein Struct. 1975, 405, 442–451. [Google Scholar]
- Wan, S.; Mak, M.W.; Kung, S.Y. Sparse regressions for predicting and interpreting subcellular localization of multi-label proteins. BMC Bioinform. 2016, 17, 97. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Mak, M.W.; Kung, S.Y. Ensemble linear neighborhood propagation for predicting subchloroplast localization of multi-location proteins. J. Proteome Res. 2016, 15, 4755–4762. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guo, M. A review of metrics measuring dissimilarity for rooted phylogenetic networks. Brief. Bioinform. 2018. [Google Scholar] [CrossRef]
- Kabli, F.; Hamou, R.M.; Amine, A. Protein classification using n-gram technique and association rules. Int. J. Softw. Innov. 2018, 6, 77. [Google Scholar] [CrossRef]
- Vries, J.K.; Liu, X. Subfamily specific conservation profiles for proteins based on n-gram patterns. BMC Bioinform. 2008, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Grama, I.; Kervrann, C.; Liu, Q. Nonlocal means and optimal weights for noise removal. SIAM J. Imaging Sci. 2017, 10, 1878–1920. [Google Scholar] [CrossRef]
- Du, P.; Gu, S.; Jiao, Y. Pseaac-general: Fast building various modes of general form of chou’s pseudo-amino acid composition for large-scale protein datasets. Int. J. Mol. Sci. 2014, 15, 3495–3506. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Wang, X.; Xu, C.; Gao, Y. Pseaac-builder: A cross-platform stand-alone program for generating various special chou’s pseudo-amino acid compositions. Anal. Biochem. 2012, 425, 117–119. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Mak, M.W.; Kung, S.Y. Transductive learning for multi-label protein subchloroplast localization prediction. IEEE/ACM Trans. Comput. Boil. Bioinform. 2016, 14, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Kurgan, L.; Cios, K.; Chen, K. Scpred: Accurate prediction of protein structural class for sequences of twilight-zone similarity with predicting sequences. BMC Bioinform. 2008, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Jia, C. A high-accuracy protein structural class prediction algorithm using predicted secondary structural information. J. Theor. Biol. 2010, 267, 272–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ding, S.; Wang, T. High-accuracy prediction of protein structural class for low-similarity sequences based on predicted secondary structure. Biochimie 2011, 93, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ying, Z.; Ji, Q.; Liu, X.; Yi, J.; Ke, C.; Quan, Z. Hierarchical classification of protein folds using a novel ensemble classifier. PLoS ONE 2013, 8, e56499. [Google Scholar]
- Song, L.; Li, D.; Zeng, X.; Wu, Y.; Guo, L.; Zou, Q. Ndna-prot: Identification of DNA-binding proteins based on unbalanced classification. BMC Bioinformatics 2014, 15, 298. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Liao, M.; Gao, X.; Zou, Q. Enhanced protein fold prediction method through a novel feature extraction technique. IEEE T. Nanobiosci. 2015, 14, 649. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Tang, J.; Zou, Q. Local-dpp: An improved DNA-binding protein prediction method by exploring local evolutionary information. Inform. Sciences 2016, 384. [Google Scholar] [CrossRef]
Sample Availability: Samples of the test dataset are available from the authors. |

| Feature Vector | Kernel Function | Acc | Sens | Spec | F-score | Mcc |
|---|---|---|---|---|---|---|
| Fn-gram | linear | 80.836% | 85.576% | 76.174% | 81.579% | 61.996% |
| FPseAAC | linear | 81.259% | 81.677% | 80.836% | 81.210% | 62.532% |
| Gaussian | 84.882% | 84.192% | 85.560% | 84.665% | 69.766% | |
| Fpssm-20 | linear | 80.402% | 76.262% | 85.760% | 79.863% | 62.378% |
| Gaussian | 76.823% | 62.769% | 88.829% | 72.222% | 53.404% | |
| Fpssm-380 | linear | 82.832% | 75.378% | 88.886% | 80.836% | 64.865% |
| Gaussian | 77.588% | 63.533% | 93.250% | 74.470% | 59.732% | |
| Fsss | linear | 74.242% | 77.585% | 70.954% | 74.939% | 48.636% |
| Gaussian | 72.950% | 72.616% | 73.296% | 72.716% | 45.912% |
| Feature Vector | Kernel Function | Acc | Sens | Spec | F-Score | Mcc |
|---|---|---|---|---|---|---|
| Fsp | linear | 84.081% | 84.373% | 83.797% | 84.025% | 68.167% |
| Gaussian | 86.149% | 85.412% | 86.885% | 85.957% | 72.299% | |
| Fpssm | linear | 85.700% | 81.505% | 89.836% | 84.970% | 71.621% |
| Gaussian | 78.526% | 62.766% | 94.050% | 74.346% | 59.913% | |
| Fpsp | linear | 89.722% | 88.284% | 91.132% | 89.506% | 79.473% |
| Gaussian | 86.969% | 86.046% | 87.882% | 86.765% | 73.951% | |
| Fpspn | linear | 89.923% | 88.492% | 91.325% | 89.706% | 79.870% |
| Gaussian | 85.531% | 84.951% | 86.110% | 85.358% | 71.062% |
| C = 2−5 | C = 2−4 | C = 2−3 | C = 2−2 | C = 2−1 | C = 20 | C = 21 | C = 22 | C = 23 | C = 24 | C = 25 | |
| γ = 2−5 | 83.936 | 84.726 | 85.769 | 86.934 | 88.285 | 89.244 | 89.974 | 90.451 | 90.252 | 89.878 | 89.558 |
| γ = 2−4 | 83.604 | 84.316 | 85.148 | 86.451 | 87.754 | 88.912 | 89.558 | 89.606 | 89.491 | 89.371 | 89.286 |
| γ = 2−3 | 76.624 | 79.972 | 82.856 | 85.962 | 86.626 | 87.917 | 88.388 | 88.484 | 88.466 | 88.472 | 88.472 |
| γ = 2−2 | 56.476 | 61.983 | 64.523 | 68.565 | 73.578 | 80.817 | 81.402 | 81.378 | 81.378 | 81.378 | 81.378 |
| γ = 2−1 | 50.407 | 50.407 | 50.407 | 50.570 | 56.995 | 67.696 | 69.259 | 69.259 | 69.259 | 69.259 | 69.259 |
| γ = 20 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 51.433 | 52.404 | 52.404 | 52.404 | 52.404 | 52.404 |
| γ = 21 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.419 | 50.413 | 50.413 | 50.413 | 50.413 | 50.413 |
| γ = 22 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 |
| γ = 23 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 |
| γ = 24 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 |
| γ = 25 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 | 50.407 |
| Times | Acc | Sens | Spec | F-score | Mcc |
|---|---|---|---|---|---|
| 1 | 90.748% ± 0.567% | 89.181% ± 1.003% | 92.287% ± 0.880% | 90.538% ± 0.617% | 81.533% ± 1.133% |
| 2 | 90.965% ± 0.348% | 89.297% ± 1.090% | 92.632% ± 0.650% | 90.750% ± 0.375% | 81.980% ± 0.657% |
| 3 | 90.851% ± 0.620% | 89.238% ± 0.706% | 92.450% ± 0.836% | 90.644% ± 0.558% | 81.737% ± 1.235% |
| 4 | 90.954% ± 0.695% | 89.343% ± 0.890% | 92.539% ± 0.727% | 90.743% ± 0.772% | 81.942% ± 1.395% |
| 5 | 90.775% ± 0.646% | 89.264% ± 1.327% | 92.267% ± 0.804% | 90.571% ± 0.689% | 81.591% ± 1.269% |
| 6 | 90.819% ± 0.546% | 89.232% ± 0.776% | 92.382% ± 1.097% | 90.612% ± 0.501% | 81.673% ± 1.101% |
| 7 | 90.813% ± 0.682% | 89.120% ± 1.024% | 92.462% ± 0.960% | 90.592% ± 0.725% | 81.660% ± 1.367% |
| 8 | 90.868% ± 0.619% | 89.232% ± 0.964% | 92.473% ± 0.580% | 90.649% ± 0.727% | 81.766% ± 1.238% |
| 9 | 90.895% ± 0.580% | 89.302% ± 0.752% | 92.460% ± 0.863% | 90.692% ± 0.458% | 81.816% ± 1.151% |
| 10 | 90.688% ± 0.532% | 89.134% ± 0.985% | 92.224% ± 0.567% | 90.478% ± 0.572% | 81.407% ± 1.048% |
| Feature Vector | Kernel Function | Acc | Sens | Spec | F-Score | Mcc |
|---|---|---|---|---|---|---|
| FPseAAC | Gaussian | 92.315% | 31.948% | 98.946% | 44.943% | 46.357% |
| Fsp | Gaussian | 92.875% | 37.385% | 98.963% | 50.743% | 51.529% |
| Fpssm-380 | linear | 93.520% | 43.640% | 98.826% | 57.745% | 57.433% |
| Fpssm | linear | 93.943% | 48.157% | 98.781% | 60.930% | 60.319% |
| Fpsp | linear | 94.980% | 62.231% | 98.572% | 70.899% | 69.132% |
| Fpspn | linear | 94.966% | 62.039% | 98.574% | 70.782% | 68.989% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Wang, J.; Zheng, Z.; Bai, X. A New Method for Recognizing Cytokines Based on Feature Combination and a Support Vector Machine Classifier. Molecules 2018, 23, 2008. https://doi.org/10.3390/molecules23082008
Yang Z, Wang J, Zheng Z, Bai X. A New Method for Recognizing Cytokines Based on Feature Combination and a Support Vector Machine Classifier. Molecules. 2018; 23(8):2008. https://doi.org/10.3390/molecules23082008
Chicago/Turabian StyleYang, Zhe, Juan Wang, Zhida Zheng, and Xin Bai. 2018. "A New Method for Recognizing Cytokines Based on Feature Combination and a Support Vector Machine Classifier" Molecules 23, no. 8: 2008. https://doi.org/10.3390/molecules23082008





