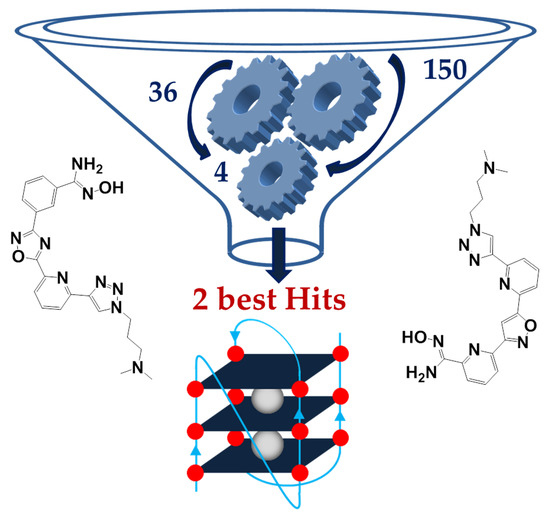
A Fragment-Based Approach for the Development of G-Quadruplex Ligands: Role of the Amidoxime Moiety
Abstract
:1. Introduction
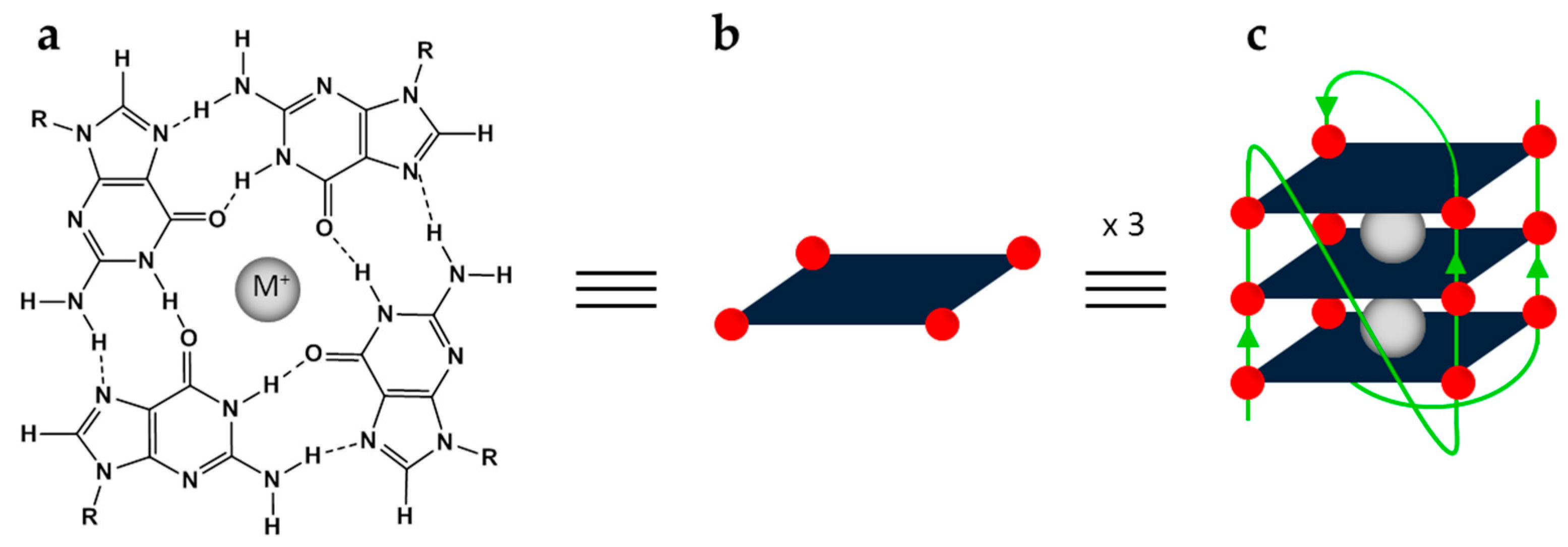
2. Results and Discussion
2.1. Initial Screening of Low-Molecular Weight Fragment Molecules
2.2. Design, Synthesis and Analysis of the Polyheteroaryl Oxadiazole/Pyridine-Ligands
2.3. Design, Synthesis and Analysis of the Final Lead Compounds
3. Conclusions
4. Materials and Methods
4.1. General Information
4.2. Synthetic Methods
4.2.1. Synthesis of the Fragment Family Shown in Figure 2a
4.2.2. Synthesis of the Fragment Family Shown in Figure 2b
4.2.3. Synthesis of the Fragment Family Shown in Figure 2c
4.3. Biophysical and Biological Assays
4.3.1. FRET Assay
4.3.2. Circular Dichroism (CD)
4.3.3. Taq Polymerase Stop Assay
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lipps, H.J.; Rhodes, D. G-quadruplex structures: In vivo evidence and function. Trends Cell Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Brosh, R.M., Jr. G-quadruplex nucleic acids and human disease. FEBS J. 2010, 277, 3470–3488. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.H.; Neidle, S. G-quadruplexes and metal ions. Met. Ions Life Sci. 2012, 10, 119–134. [Google Scholar] [PubMed]
- Sen, D.; Gilbert, W. A sodium-potassium switch in the formation of four-stranded g4-DNA. Nature 1990, 344, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.P. Neurodegenerative diseases: G-quadruplex poses quadruple threat. Nature 2014, 507, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Cogoi, S.; Xodo, L.E. G-quadruplex formation within the promoter of the kras proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006, 34, 2536–2549. [Google Scholar] [CrossRef] [PubMed]
- Eddy, J.; Maizels, N. Gene function correlates with potential for g4 DNA formation in the human genome. Nucleic Acids Res. 2006, 34, 3887–3896. [Google Scholar] [CrossRef] [PubMed]
- Eddy, J.; Maizels, N. Conserved elements with potential to form polymorphic g-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008, 36, 1321–1333. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Nakken, S.; Rognes, T.; Hovig, E. The disruptive positions in human g-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 2009, 37, 5749–5756. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Yadav, V.K.; Basundra, R.; Kumar, A.; Chowdhury, S. Evidence of genome-wide g4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Res. 2009, 37, 4194–4204. [Google Scholar] [CrossRef] [PubMed]
- Mani, P.; Yadav, V.K.; Das, S.K.; Chowdhury, S. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS ONE 2009, 4, e4399. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.W.; Pereira, F.; Barrett, S.P.; Kolesar, J.E.; Cao, K.; Damas, J.; Yatsunyk, L.A.; Johnson, F.B.; Kaufman, B.A. Association of g-quadruplex forming sequences with human mtdna deletion breakpoints. BMC Genom. 2014, 15, 677. [Google Scholar] [CrossRef] [PubMed]
- Nambiar, M.; Srivastava, M.; Gopalakrishnan, V.; Sankaran, S.K.; Raghavan, S.C. G-quadruplex structures formed at the hox11 breakpoint region contribute to its fragility during t(10;14) translocation in t-cell leukemia. Mol. Cell Biol. 2013, 33, 4266–4281. [Google Scholar] [CrossRef] [PubMed]
- Sissi, C.; Gatto, B.; Palumbo, M. The evolving world of protein-g-quadruplex recognition: A medicinal chemist’s perspective. Biochimie 2011, 93, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Scalabrin, M.; Frasson, I.; Ruggiero, E.; Perrone, R.; Tosoni, E.; Lago, S.; Tassinari, M.; Palu, G.; Richter, S.N. The cellular protein hnrnp a2/b1 enhances hiv-1 transcription by unfolding ltr promoter g-quadruplexes. Sci. Rep. 2017, 7, 45244. [Google Scholar] [CrossRef] [PubMed]
- Tosoni, E.; Frasson, I.; Scalabrin, M.; Perrone, R.; Butovskaya, E.; Nadai, M.; Palu, G.; Fabris, D.; Richter, S.N. Nucleolin stabilizes g-quadruplex structures folded by the ltr promoter and silences hiv-1 viral transcription. Nucleic Acids Res. 2015, 43, 8884–8897. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA g-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M., Jr.; Sen, D.; Lansdorp, P.M. Detection of g-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; Miller, J.; Howat, W.J.; Balasubramanian, S. Elevated levels of g-quadruplex formation in human stomach and liver cancer tissues. PLoS ONE 2014, 9, e102711. [Google Scholar] [CrossRef] [PubMed]
- Seifert, H.S. Above and beyond watson and crick: Guanine quadruplex structures and microbes. Annu. Rev. Microbiol. 2018, 72. [Google Scholar] [CrossRef] [PubMed]
- Artusi, S.; Nadai, M.; Perrone, R.; Biasolo, M.A.; Palu, G.; Flamand, L.; Calistri, A.; Richter, S.N. The herpes simplex virus-1 genome contains multiple clusters of repeated g-quadruplex: Implications for the antiviral activity of a g-quadruplex ligand. Antiviral Res. 2015, 118, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting g-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Perrone, R.; Butovskaya, E.; Daelemans, D.; Palu, G.; Pannecouque, C.; Richter, S.N. Anti-hiv-1 activity of the g-quadruplex ligand braco-19. J. Antimicrob. Chemother. 2014, 69, 3248–3258. [Google Scholar] [CrossRef] [PubMed]
- Perrone, R.; Nadai, M.; Frasson, I.; Poe, J.A.; Butovskaya, E.; Smithgall, T.E.; Palumbo, M.; Palu, G.; Richter, S.N. A dynamic g-quadruplex region regulates the hiv-1 long terminal repeat promoter. J. Med. Chem. 2013, 56, 6521–6530. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, E.; Richter, S.N. G-quadruplexes and g-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef] [PubMed]
- Hajduk, P.J.; Greer, J. A decade of fragment-based drug design: Strategic advances and lessons learned. Nat. Rev. Drug Discov. 2007, 6, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Congreve, M.; Carr, R.; Murray, C.; Jhoti, H. A ‘rule of three’ for fragment-based lead discovery? Drug Discov. Today 2003, 8, 876–877. [Google Scholar] [CrossRef]
- Baker, M. Fragment-based lead discovery grows up. Nat. Rev. Drug Discov. 2013, 12, 5–7. [Google Scholar] [CrossRef] [PubMed]
- De Kloe, G.E.; Bailey, D.; Leurs, R.; de Esch, I.J. Transforming fragments into candidates: Small becomes big in medicinal chemistry. Drug Discov. Today 2009, 14, 630–646. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, M.; Law, R.J.; Ichihara, O.; Hesterkamp, T.; Hallett, D. Fragments: Past, present and future. Drug Discov. Today Technol. 2010, 7, 147–202. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.; Lee, J.T.; Wang, W.; Zhang, J.; Cho, H.; Mamo, S.; Bremer, R.; Gillette, S.; Kong, J.; Haass, N.K.; et al. Discovery of a selective inhibitor of oncogenic b-raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. USA 2008, 105, 3041–3046. [Google Scholar] [CrossRef] [PubMed]
- Nasiri, H.R.; Bell, N.M.; McLuckie, K.I.; Husby, J.; Abell, C.; Neidle, S.; Balasubramanian, S. Targeting a c-myc g-quadruplex DNA with a fragment library. Chem. Commun. 2014, 50, 1704–1707. [Google Scholar] [CrossRef] [PubMed]
- Di Antonio, M.; Biffi, G.; Mariani, A.; Raiber, E.A.; Rodriguez, R.; Balasubramanian, S. Selective rna versus DNA g-quadruplex targeting by in situ click chemistry. Angew. Chem. Int. Ed. Engl. 2012, 51, 11073–11078. [Google Scholar] [CrossRef] [PubMed]
- Perrone, R.; Doria, F.; Butovskaya, E.; Frasson, I.; Botti, S.; Scalabrin, M.; Lago, S.; Grande, V.; Nadai, M.; Freccero, M.; et al. Synthesis, binding and antiviral properties of potent core-extended naphthalene diimides targeting the hiv-1 long terminal repeat promoter g-quadruplexes. J. Med. Chem. 2015, 58, 9639–9652. [Google Scholar] [CrossRef] [PubMed]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (ttaggg)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef] [PubMed]
- Renciuk, D.; Zhou, J.; Beaurepaire, L.; Guedin, A.; Bourdoncle, A.; Mergny, J.L. A fret-based screening assay for nucleic acid ligands. Methods (San Diego, Calif.) 2012, 57, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Havemeyer, A.; Bittner, F.; Wollers, S.; Mendel, R.; Kunze, T.; Clement, B. Identification of the missing component in the mitochondrial benzamidoxime prodrug-converting system as a novel molybdenum enzyme. J. Biol. Chem. 2006, 281, 34796–34802. [Google Scholar] [CrossRef] [PubMed]
- Fylaktakidou, K.C.; Hadjipavlou-Litina, D.J.; Litinas, K.E.; Varella, E.A.; Nicolaides, D.N. Recent developments in the chemistry and in the biological applications of amidoximes. Curr. Pharm. Des. 2008, 14, 1001–1047. [Google Scholar] [CrossRef] [PubMed]
- Clement, B.; Raether, W. Amidoximes of pentamidine: Synthesis, trypanocidal and leishmanicidal activity. Arzneimittelforschung 1985, 35, 1009–1014. [Google Scholar] [PubMed]
- Petenzi, M.; Verga, D.; Largy, E.; Hamon, F.; Doria, F.; Teulade-Fichou, M.P.; Guedin, A.; Mergny, J.L.; Mella, M.; Freccero, M. Cationic pentaheteroaryls as selective g-quadruplex ligands by solvent-free microwave-assisted synthesis. Chemistry 2012, 18, 14487–14496. [Google Scholar] [CrossRef] [PubMed]
- Doria, F.; Manet, I.; Grande, V.; Monti, S.; Freccero, M. Water-soluble naphthalene diimides as singlet oxygen sensitizers. J. Org. Chem. 2013, 78, 8065–8073. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Sanders, D.A.; Di Antonio, M.; Matsis, S.; Riou, J.F.; Rodriguez, R.; Balasubramanian, S. Pyridostatin analogues promote telomere dysfunction and long-term growth inhibition in human cancer cells. Org. Biomol. Chem. 2012, 10, 6537–6546. [Google Scholar] [CrossRef] [PubMed]
- Bruton, E.A.; Brammer, L.; Christopher Pigge, F.; Aakeroy, C.B.; Leinen, D.S. Hydrogen bond patterns in aromatic and aliphatic dioximes. New J. Chem. 2003, 27, 1084–1094. [Google Scholar] [CrossRef]
- Stemper, J.; Tuo, W.; Mazarío, E.; Helal, A.S.; Djurovic, A.; Lion, C.; El Hage Chahine, J.-M.; Maurel, F.; Hémadi, M.; Le Gall, T. Synthesis of bis(amidoxime)s and evaluation of their properties as uranyl-complexing agents. Tetrahedron 2018, 74, 2641–2649. [Google Scholar] [CrossRef]
- Rachwal, P.A.; Fox, K.R. Quadruplex melting. Methods 2007, 43, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |






| Fragment | Structural Complexity | ΔTm 1 ± s.d. 2 (°C) | ||
|---|---|---|---|---|
| LTR-III | hTel | dsDNA | ||
| 1 | a | <1 | <1 | <1 |
| 2 | <1 | <1 | <1 | |
| 3 | <1 | <1 | <1 | |
| 4 | <1 | <1 | <1 | |
| 5 | 1.5 ± 0.1 | 1.0 ± 0.1 | <1 | |
| 6 | <1 | <1 | <1 | |
| 7 | <1 | <1 | <1 | |
| 8 | <1 | <1 | <1 | |
| 9 | <1 | <1 | <1 | |
| 10 | <1 | <1 | <1 | |
| 11 | 1.5 ± 0.1 | 1.5 ± 0.1 | <1 | |
| 12 | 1.5 ± 0.1 | 1.5 ± 0.1 | <1 | |
| 13 | <1 | <1 | <1 | |
| 14 | 6.0 ± 1.5 | 4.5 ± 2.2 | 2.5 ± 0.5 | |
| 15 | 1.5 ± 0.1 | 3.0 ± 0.5 | <1 | |
| 16 | b | 3.6 ± 0.6 | 2.5 ± 0.1 | <1 |
| 17 | 1.5 ± 0.1 | 3.0 ± 0.5 | <1 | |
| 18 | <1 | <1 | <1 | |
| 19 | n.d. 3 | n.d. 3 | n.d. 3 | |
| 20 | 5.5 ± 0.5 | 5.5 ± 0.5 | <1 | |
| 21 | n.d. 3 | n.d. 3 | n.d. 3 | |
| 22 | 5.5 ± 0.5 | 4.0 ± 0.5 | <1 | |
| 23 | 4.1 ± 1.3 | 2.7 ± 1.1 | <1 | |
| 24 | 7.1 ± 0.1 | 6.5 ± 0.8 | 1.1 ± 0.2 | |
| 25 | 4.5 ± 0.5 | 2.5 ± 0.1 | <1 | |
| 26 | 6.5 ± 0.5 | 5.0 ± 0.5 | <1 | |
| 27 | 13.0 ± 0.6 | 14.0 ±0.7 | <1 | |
| 28 | 6.0 ± 0.5 | 4.5 ± 0.5 | <1 | |
| 29 | 3.0 ± 0.6 | 5.4 ± 0.9 | 1.1 ± 0.2 | |
| 30 | 10.8 ± 1.0 | 14.7 ± 1.1 | <1 | |
| 31 | n.d. 3 | n.d. 3 | n.d. 3 | |
| 32 | 12.8 ± 2.0 | 11.8 ± 1.9 | <1 | |
| 33 | c | >22.1 | >24.1 | >28.0 |
| 34 | >22.1 | >24.1 | >28.0 | |
| 35 | >22.1 | >24.1 | 4.1 ± 0.2 | |
| 36 | >22.1 | >24.1 | 14.1 ± 1.0 | |
| Fragment | Structural Complexity | ΔTm 1 ± s.d. 2 (°C) | ||
|---|---|---|---|---|
| LTR-III | hTel | dsDNA | ||
| (16) | b | <1 | <1 | <1 |
| (23) | <1 | <1 | <1 | |
| (24) | 2.9 ± 0.3 | 1.1 ± 0.1 | <1 | |
| (29) | <1 | <1 | <1 | |
| (30) | 3.5 ± 0.5 | 3.0 ± 0.5 | <1 | |
| (32) | 5.3 ± 0.5 | 3.5 ± 0.4 | <1 | |
| (33) | c | 15.5 ± 0.5 | 15.3 ± 0.3 | <1 |
| (34) | 14.3 ± 0.3 | 14.0 ± 1.0 | <1 | |
| (35) | 9.0 ± 0.2 | 8.9 ± 0.1 | <1 | |
| (36) | 14.1 ± 0.2 | 14.6 ± 0.3 | <1 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tassinari, M.; Lena, A.; Butovskaya, E.; Pirota, V.; Nadai, M.; Freccero, M.; Doria, F.; Richter, S.N. A Fragment-Based Approach for the Development of G-Quadruplex Ligands: Role of the Amidoxime Moiety. Molecules 2018, 23, 1874. https://doi.org/10.3390/molecules23081874
Tassinari M, Lena A, Butovskaya E, Pirota V, Nadai M, Freccero M, Doria F, Richter SN. A Fragment-Based Approach for the Development of G-Quadruplex Ligands: Role of the Amidoxime Moiety. Molecules. 2018; 23(8):1874. https://doi.org/10.3390/molecules23081874
Chicago/Turabian StyleTassinari, Martina, Alberto Lena, Elena Butovskaya, Valentina Pirota, Matteo Nadai, Mauro Freccero, Filippo Doria, and Sara N. Richter. 2018. "A Fragment-Based Approach for the Development of G-Quadruplex Ligands: Role of the Amidoxime Moiety" Molecules 23, no. 8: 1874. https://doi.org/10.3390/molecules23081874
APA StyleTassinari, M., Lena, A., Butovskaya, E., Pirota, V., Nadai, M., Freccero, M., Doria, F., & Richter, S. N. (2018). A Fragment-Based Approach for the Development of G-Quadruplex Ligands: Role of the Amidoxime Moiety. Molecules, 23(8), 1874. https://doi.org/10.3390/molecules23081874