The Mechanism for siRNA Transmembrane Assisted by PMAL
Abstract
:1. Introduction
2. Results
2.1. Transmembrane Process for the Naked siRNA
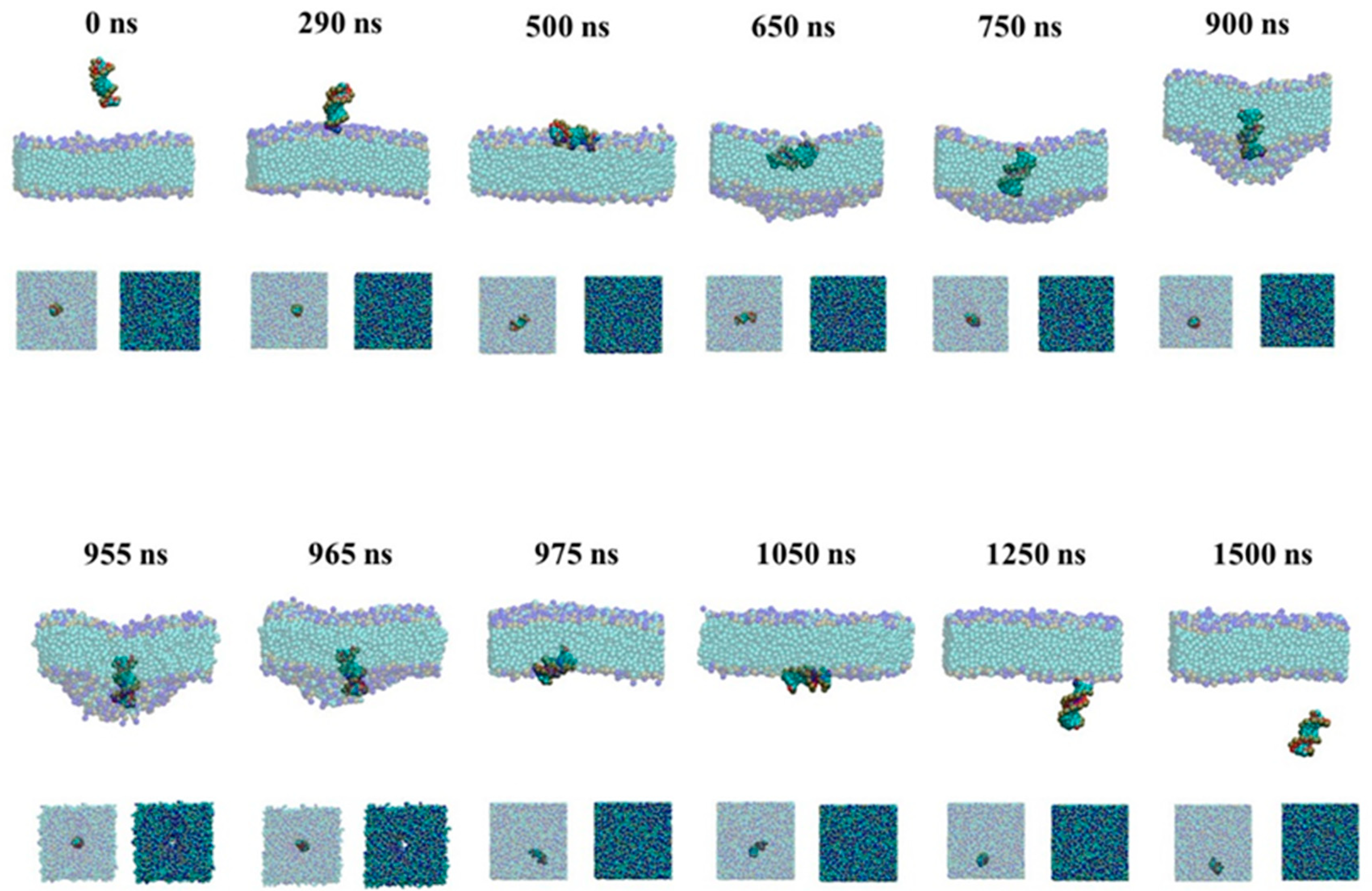
2.1.1. Active Transport Process for the Naked siRNA
2.1.2. Passive Transport Process for Single siRNA
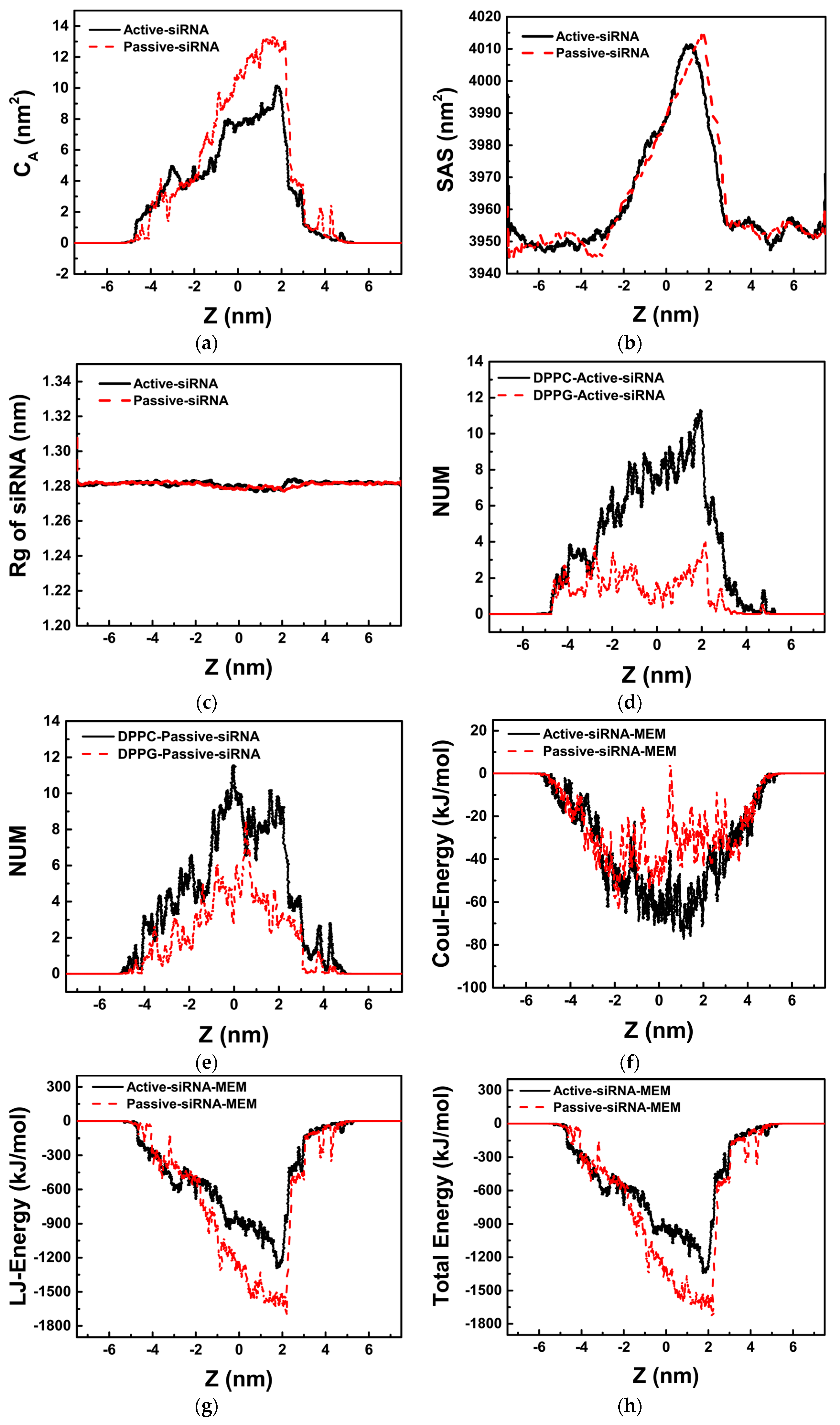
2.2. Transmembrane Process of siRNA Assisted by PMAL
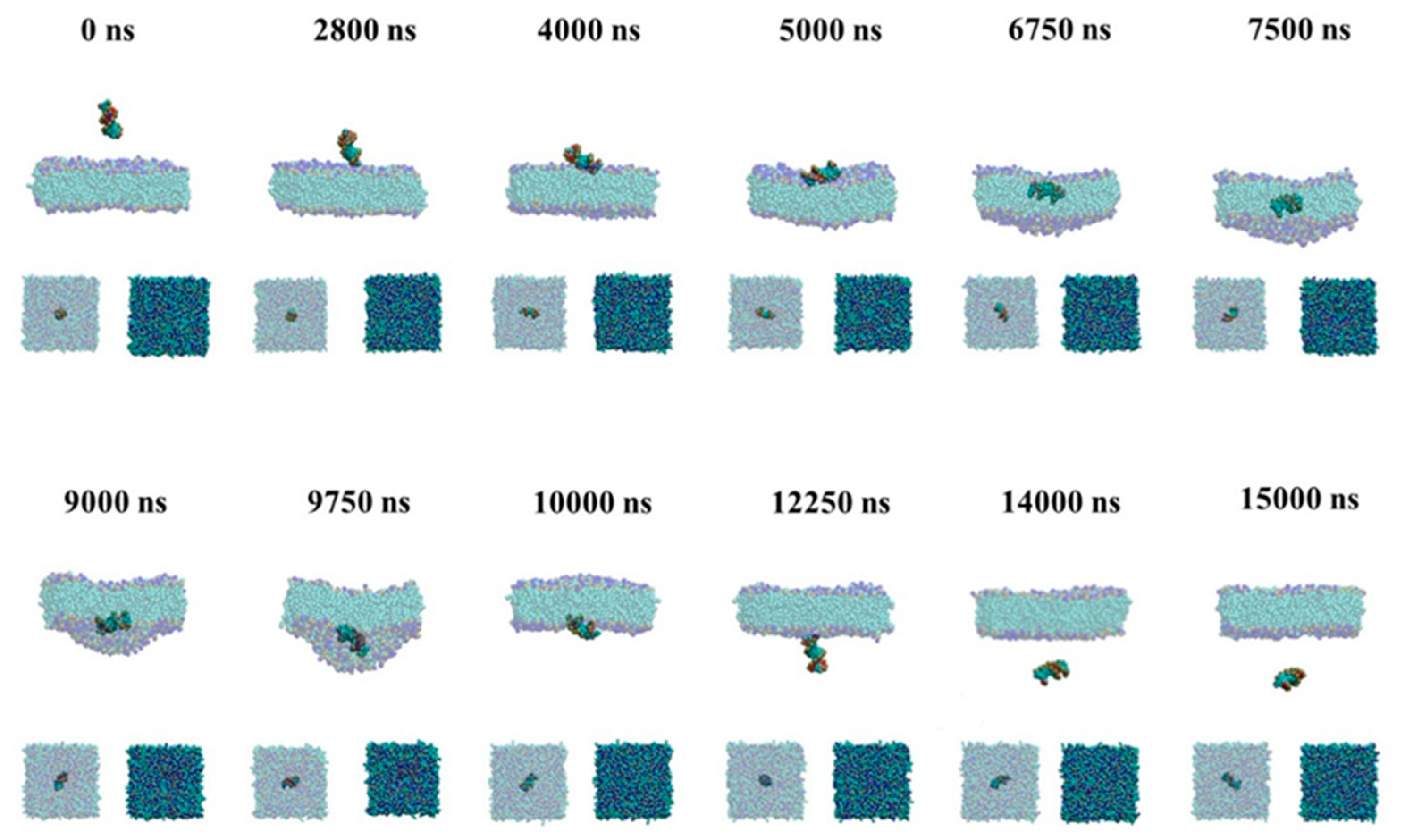
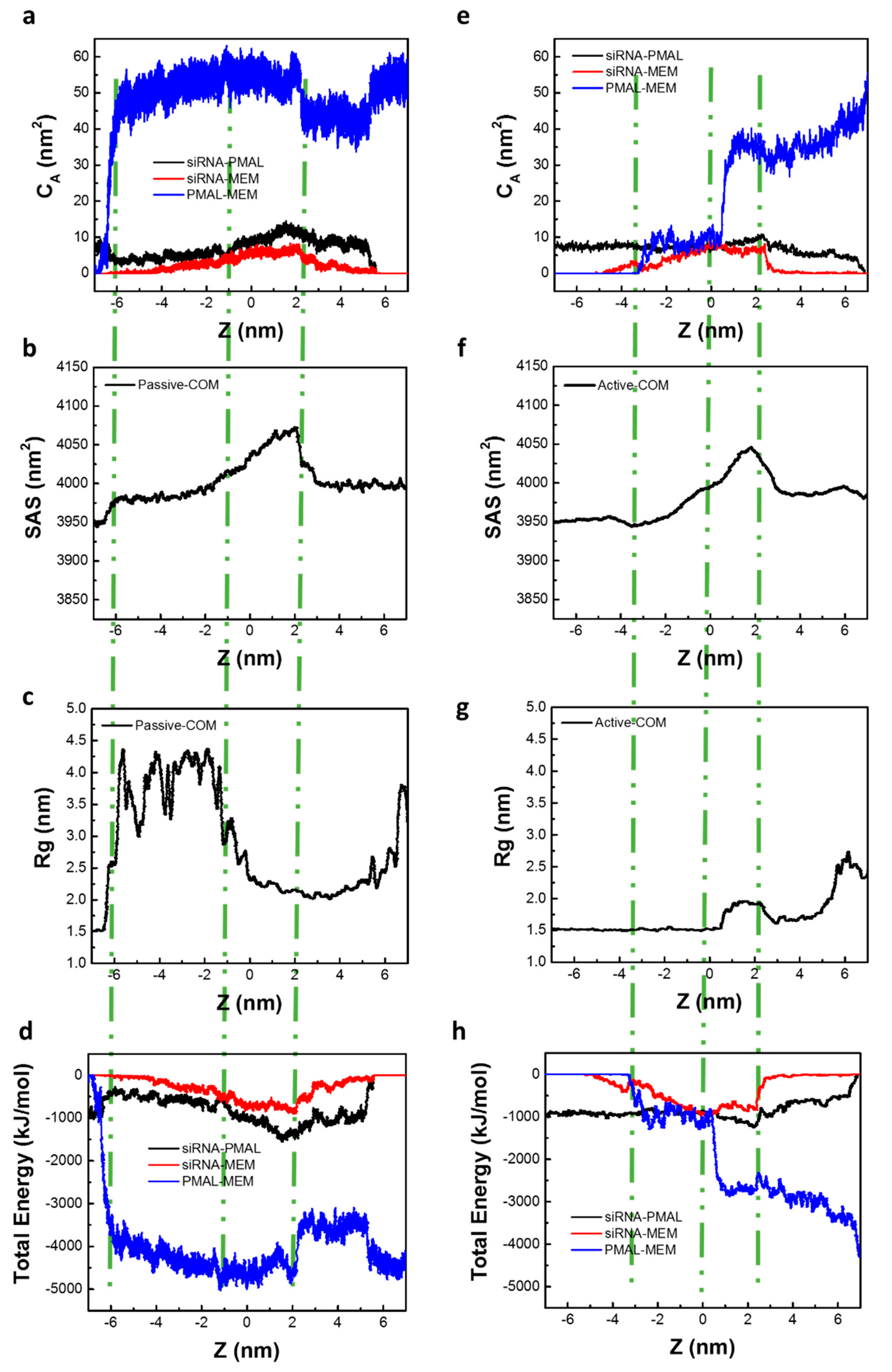
2.2.1. Passive Transport of siRNA Assisted by PMAL
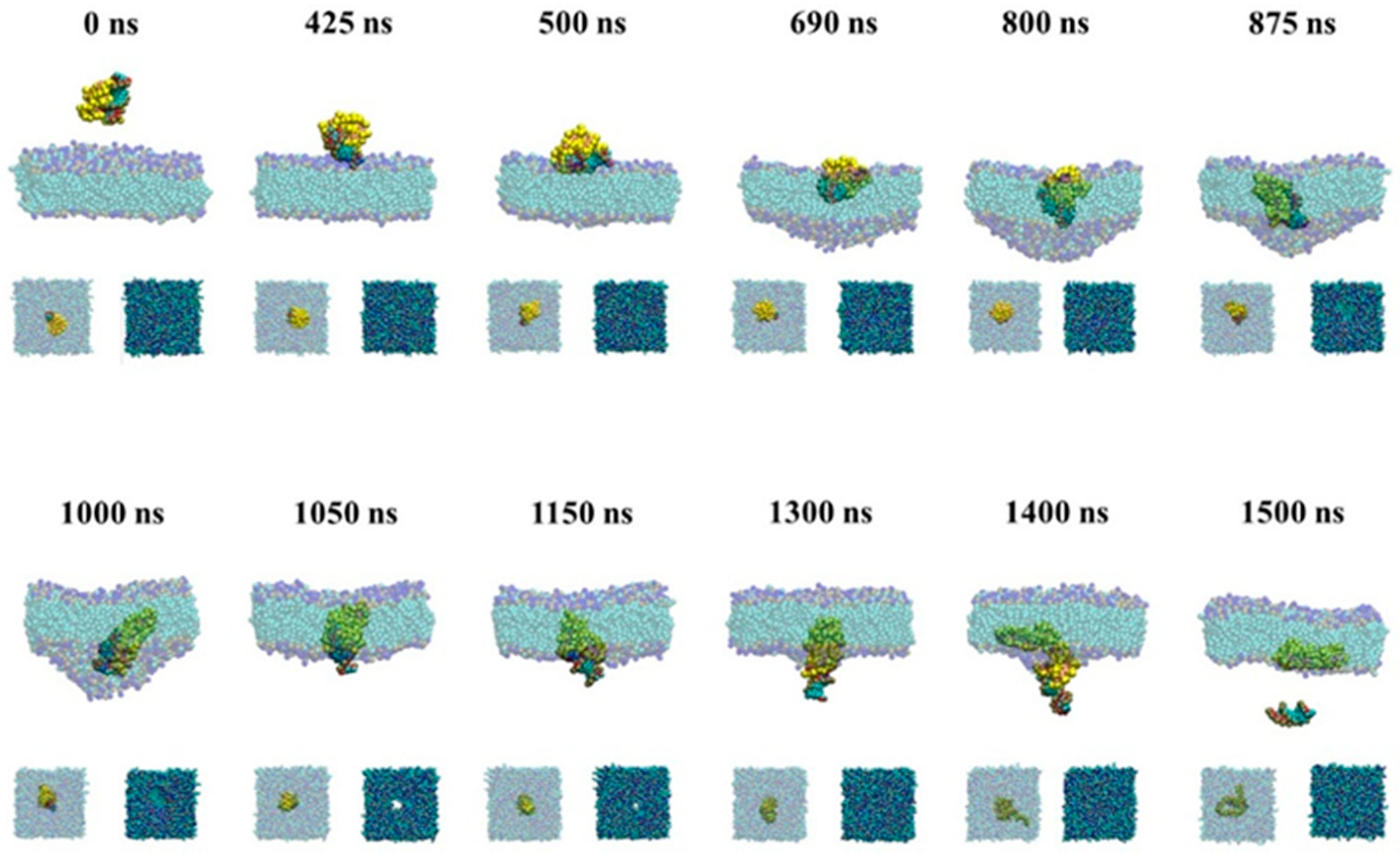
2.2.2. Active Transport of siRNA Assisted by PMAL
3. Discussion
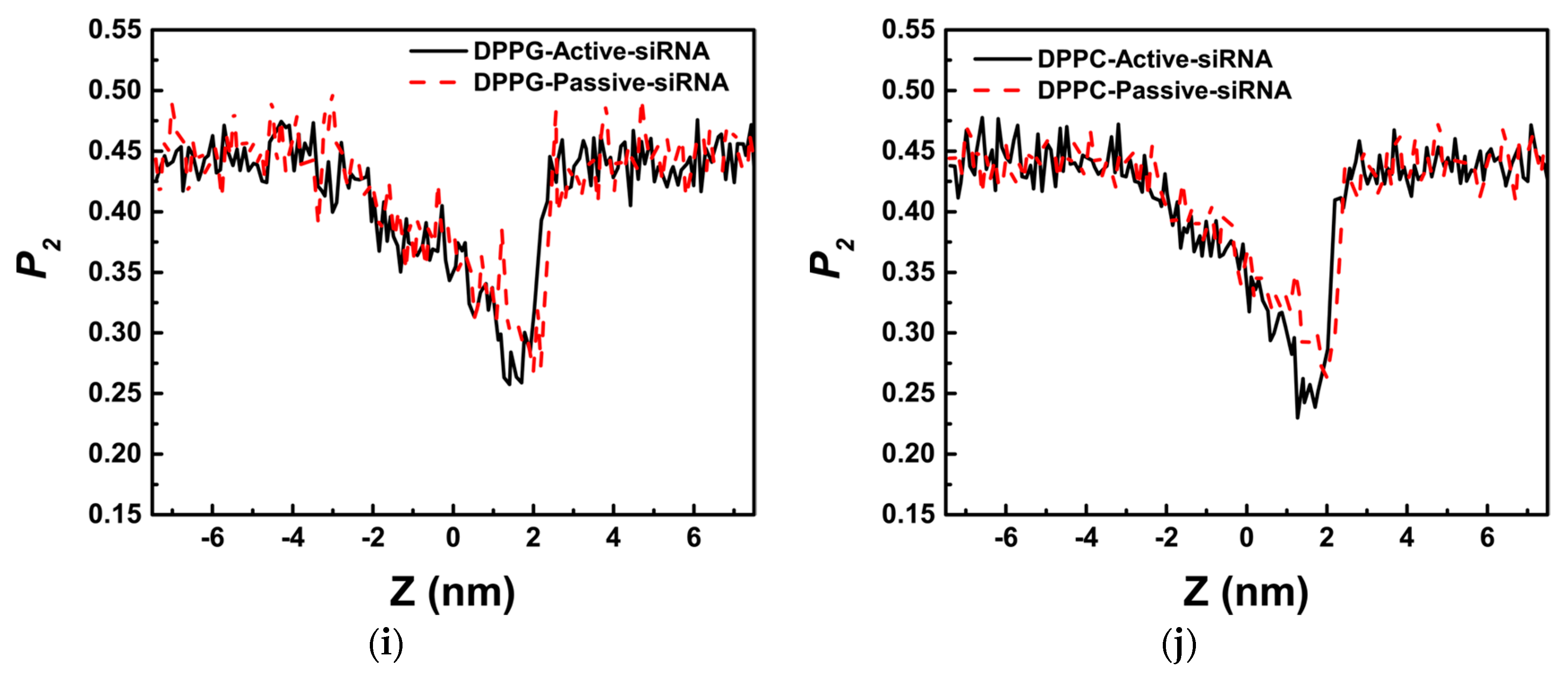
3.1. Potential of Mean Force for the Naked siRNA And siRNA-PMAL Complex During Transmembrane Process
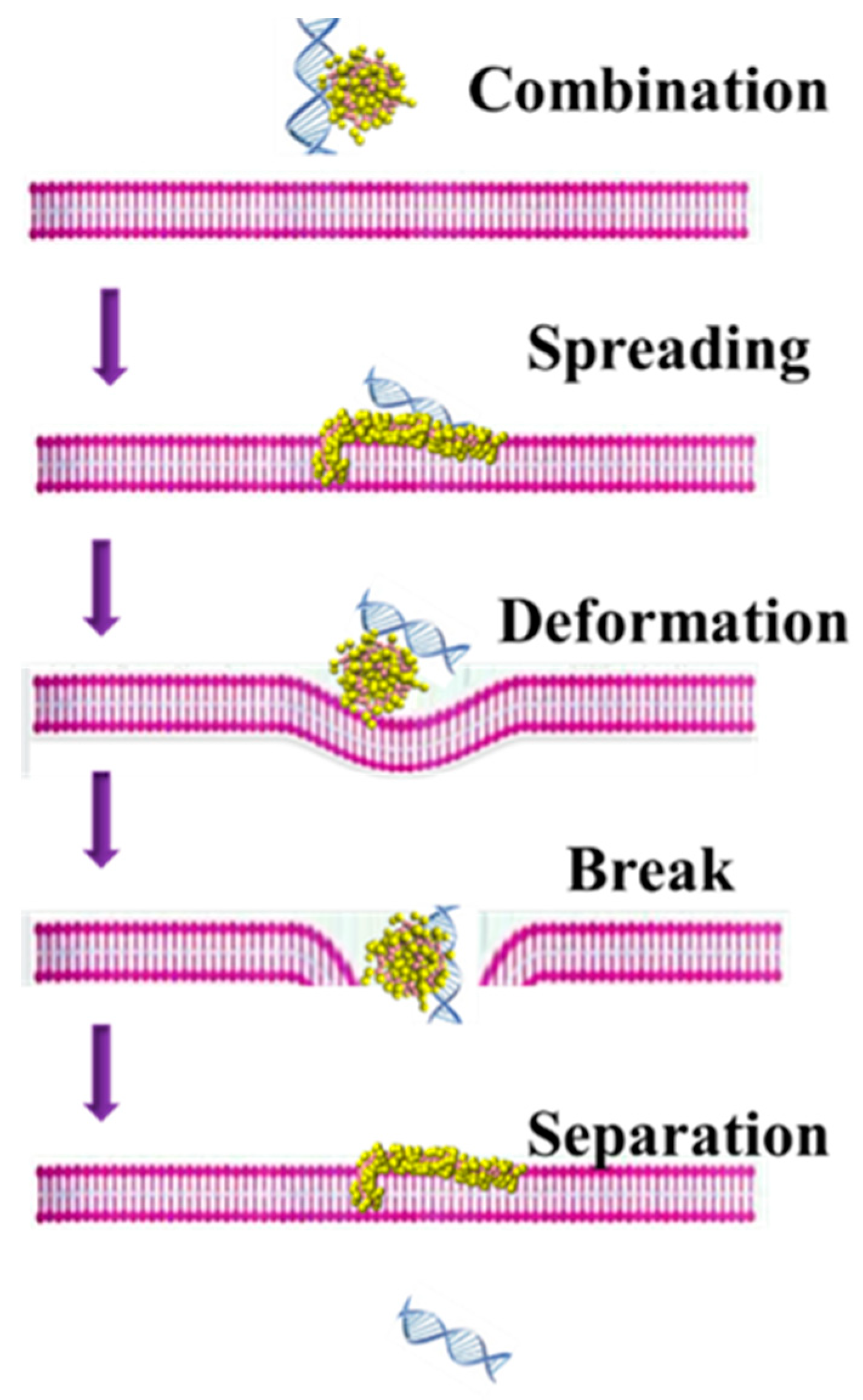
3.2. The Possible Mechanism of siRNA Delivery Assisted by PMAL
4. Materials and Methods
4.1. Models
4.1.1. Coarse-Grained Model of Membrane
4.1.2. Coarse-Grained Model of PMAL
4.1.3. Coarse-Grained Model of siRNA and Water.
4.1.4. Complex of PMAL and siRNA
4.1.5. Transmembrane Model
4.2. Simulation Details
4.2.1. Transmembrane Simulation
4.2.2. PMF Calculation
4.3. Analytical Methods
4.3.1. Radius of Gyration (Rg)
4.3.2. Contact Area (CA)
4.3.3. Order Parameter of Lipid (P2)
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Novina, C.D.; Sharp, P.A. The RNAi revolution. Nature 2004, 430, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Scherer, L.J.; Rossi, J.J. Approaches for the sequence-specific knockdown of mRNA. Nat. Biotechnol. 2003, 21, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Dykxhoorn, D.M.; Novina, C.D.; Sharp, P.A. Killing the messenger: Short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003, 4, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Bitko, V.; Musiyenko, A.; Shulyayeva, O.; Barik, S. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 2005, 11, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Schiffelers, R.M.; Ansari1, A.; Xu, J.; Zhou, Q.; Tang, Q.Q.; Storm, G.; Molema, G.; Lu, P.Y.; Scaria1, P.V.; Woodle, M.C. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004, 32, e149. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 516. [Google Scholar] [CrossRef]
- Barquinero, J.; Eixarch, H.; Perez-Melgosa, M. Retroviral vectors: New applications for an old tool. Gene Ther. 2004, 11, S3–S9. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Fisher, K.A.; Darcy, R.; Cryan, J.F.; O’Driscoll, C. Therapeutic targeting in the silent era: Advances in non-viral siRNA delivery. Mol. Biosyst. 2010, 6, 1143–1161. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.; Benter, I. Toxicogenomics of non-viral drug delivery systems for RNAi: Potential impact on siRNA-mediated gene silencing activity and specificity. Adv. Drug Deliv. Rev. 2007, 59, 164–182. [Google Scholar] [CrossRef] [PubMed]
- Grayson, A.C.R.; Doody, A.M.; Putnam, D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm. Res. 2006, 23, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, S.H.; Park, T.G. Intracellular siRNA delivery system using polyelectrolyte complex micelles prepared from VEGF siRNA-PEG conjugate and cationic fusogenic peptide. Biochem. Biophys. Res. Commun. 2007, 357, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Katas, H.; Alpar, H.O. Development and characterisation of chitosan nanoparticles for siRNA delivery. J. Control. Release 2006, 115, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Corbet, C.; Ragelle, H.; Pourcelle, V.; Vanvarenberg, K.; Marchand-Brynaert, J.; Préat, V.; Feron, O. Delivery of siRNA targeting tumor metabolism using non-covalent PEGylated chitosan nanoparticles: Identification of an optimal combination of ligand structure, linker and grafting method. J. Control. Release 2016, 223, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Davaa, E.; Myung, C.S.; Park, J.S. Enhanced siRNA delivery using cationic liposomes with new polyarginine-conjugated PEG-lipid. Int. J. Pharm. 2010, 392, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Cai, J.; Borjihan, W.; Ganbold, T.; Rana, T.M.; Baigude, H. Preparation of novel curdlan nanoparticles for intracellular siRNA delivery. Carbohydr. Polym. 2015, 117, 324–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, M.E.; Zuckerman, J.E.; Choi, C.H.J.; Seligson, D.; Tolcher, A.; Alabi, C.A.; Yen, Y.; Heidel, J.D.; Ribas, A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010, 464, 1067–1140. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.L.; Zhang, M.; Minko, T. Multifunctional triblock nanocarrier (PAMAM-PEG-PLL) for the efficient intracellular siRNA delivery and gene silencing. ACS Nano 2011, 5, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Winters, M.; Holodniy, M.; Dai, H.J. siRNA delivery into human T cells and primary cells with carbon-nanotube transporters. Angew. Chem. Int. Ed. 2007, 46, 2023–2027. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.X.; Little, H.C.; Tiambeng, T.N.; Williams, G.A.; Guan, Z.B. Multifunctional dendronized peptide polymer platform for safe and effective siRNA delivery. J. Am. Chem. Soc. 2013, 135, 4962–4965. [Google Scholar] [CrossRef] [PubMed]
- Forbes, D.C.; Peppas, N.A. Polycationic nanoparticles for siRNA delivery: Comparing ARGET ATRP and UV-initiated formulations. ACS Nano 2014, 8, 2908–2917. [Google Scholar] [CrossRef] [PubMed]
- Dahlman, J.E.; Barnes, C.; Khan, O.F.; Thiriot, A.; Jhunjunwala, S.; Shaw, T.E.; Xing, Y.; Sager, H.B.; Sahay, G.; Speciner, L. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol. 2014, 9, 648–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Ouyang, Y.; Kong, X.; Zhu, J.; Lu, D.; Liu, Z. A multi-scale molecular dynamics simulation of PMAL facilitated delivery of siRNA. RSC Adv. 2015, 5, 68227–68233. [Google Scholar] [CrossRef]
- Pombo-Garcia, K.; Weiss, S.; Zarschler, K.; Ang, C.S.; Hübner, R.; Pufe, J.; Meister, S.; Seidel, J.; Pietzsch, J.; Spiccia, L. Zwitterionic polymer-coated ultrasmall superparamagnetic iron oxide nanoparticles with low protein interaction and high biocompatibility. Chemnanomat 2016, 2, 959–971. [Google Scholar] [CrossRef]
- Xu, X.; Kanduc, M.; Wu, J.Z.; Dzubiella, J. Potential of mean force and transient states in polyelectrolyte pair complexation. J. Chem. Phys. 2016, 145, 034901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlitz, H.; Li, C.W.; Wu, C.X.; Sommer, J.U. Polyelectrolyte brushes in external fields: Molecular dynamics simulations and mean-field theory. Soft Matter 2015, 11, 5688–5696. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, J.M.Y.; Dobrynin, A.V. Polyelectrolytes in salt solutions: Molecular dynamics simulations. Macromolecules 2011, 44, 5798–5816. [Google Scholar] [CrossRef]
- Meneksedag-Erol, D.; Tang, T.; Uludag, H. Molecular modeling of polynucleotide complexes. Biomaterials 2014, 35, 7068–7076. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, D.F.; Zhang, H.; Parekh, H.S.; Smith, S.C. Structure and dynamics of multiple cationic vectors-siRNA complexation by all-atomic molecular dynamics simulations. J. Phys. Chem. B 2010, 114, 9231–9237. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Tang, T.; Uludag, H. A molecular dynamics simulation study on the effect of lipid substitution on polyethylenimine mediated siRNA complexation. Biomaterials 2013, 34, 2822–2833. [Google Scholar] [CrossRef] [PubMed]
- Karatasos, K.; Posocco, P.; Laurini, E.; Pricl, S. Poly(amidoamine)-based dendrimer/siRNA complexation studied by computer simulations: Effects of pH and generation on dendrimer structure and siRNA binding. Macromol. Biosci. 2012, 12, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.F.; Li, J.P.; Lu, D.N. The mechanism for the complexation and dissociation between siRNA and PMAL: A molecular dynamics simulation study based on a coarse-grained model. Mol. Simul. 2017, 43, 1385–1393. [Google Scholar] [CrossRef]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, A.; Beyer, K.; Neubert, R.; Garidel, P.; Blume, A. Solubilization of negatively charged dppc/dppg liposomes by bile salts. J. Colloid Interface Sci. 2004, 279, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Harishchandra, R.K.; Wulff, S.; Lentzen, G.; Neuhaus, T.; Galla, H.J. The effect of compatible solute ectoines on the structural organization of lipid monolayer and bilayer membranes. Biophys. Chem. 2010, 150, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. Model. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single-crystals—A new molecular-dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartels, C. Analyzing biased monte carlo and molecular dynamics simulations. Chem. Phys. Lett. 2000, 331, 446–454. [Google Scholar] [CrossRef]
- Kumar, S.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A.; Rosenberg, J.M. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The Method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Gabriel, B.; Teissie, J. Proton lateral conduction along lipid monolayers is present only in the liquid-expanded state. J. Am. Chem. Soc. 1991, 113, 8818–8821. [Google Scholar] [CrossRef]
- Tamm, L.K.; Mcconnell, H.M. Supported phospholipid bilayers. Biophys. J. 1985, 47, 105. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Li, J.; Su, N.; Lu, D. The Mechanism for siRNA Transmembrane Assisted by PMAL. Molecules 2018, 23, 1586. https://doi.org/10.3390/molecules23071586
Lu Y, Li J, Su N, Lu D. The Mechanism for siRNA Transmembrane Assisted by PMAL. Molecules. 2018; 23(7):1586. https://doi.org/10.3390/molecules23071586
Chicago/Turabian StyleLu, Yanfei, Jipeng Li, Nan Su, and Diannan Lu. 2018. "The Mechanism for siRNA Transmembrane Assisted by PMAL" Molecules 23, no. 7: 1586. https://doi.org/10.3390/molecules23071586
APA StyleLu, Y., Li, J., Su, N., & Lu, D. (2018). The Mechanism for siRNA Transmembrane Assisted by PMAL. Molecules, 23(7), 1586. https://doi.org/10.3390/molecules23071586




