Pterostilbene and 4′-Methoxyresveratrol Inhibited Lipopolysaccharide-Induced Inflammatory Response in RAW264.7 Macrophages
Abstract
:1. Introduction
2. Results
2.1. Effects of Pte and 4MR on Cell Viability
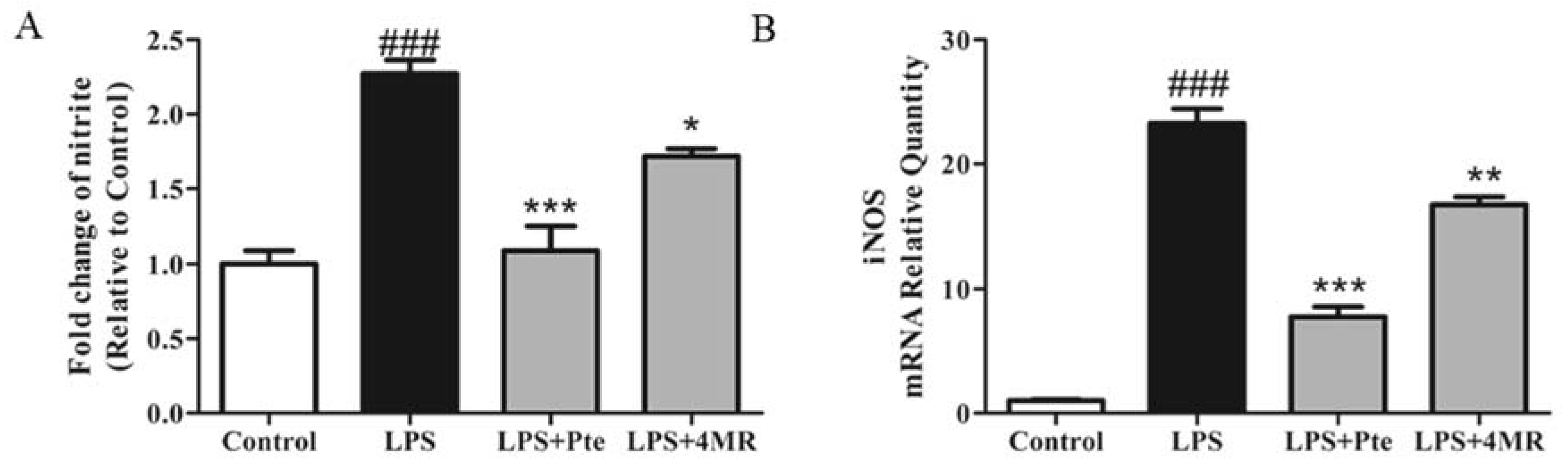
2.2. Pte and 4MR Inhibited LPS-Induced NO Production by Attenuating iNOS Expression in RAW 264.7 Macrophages
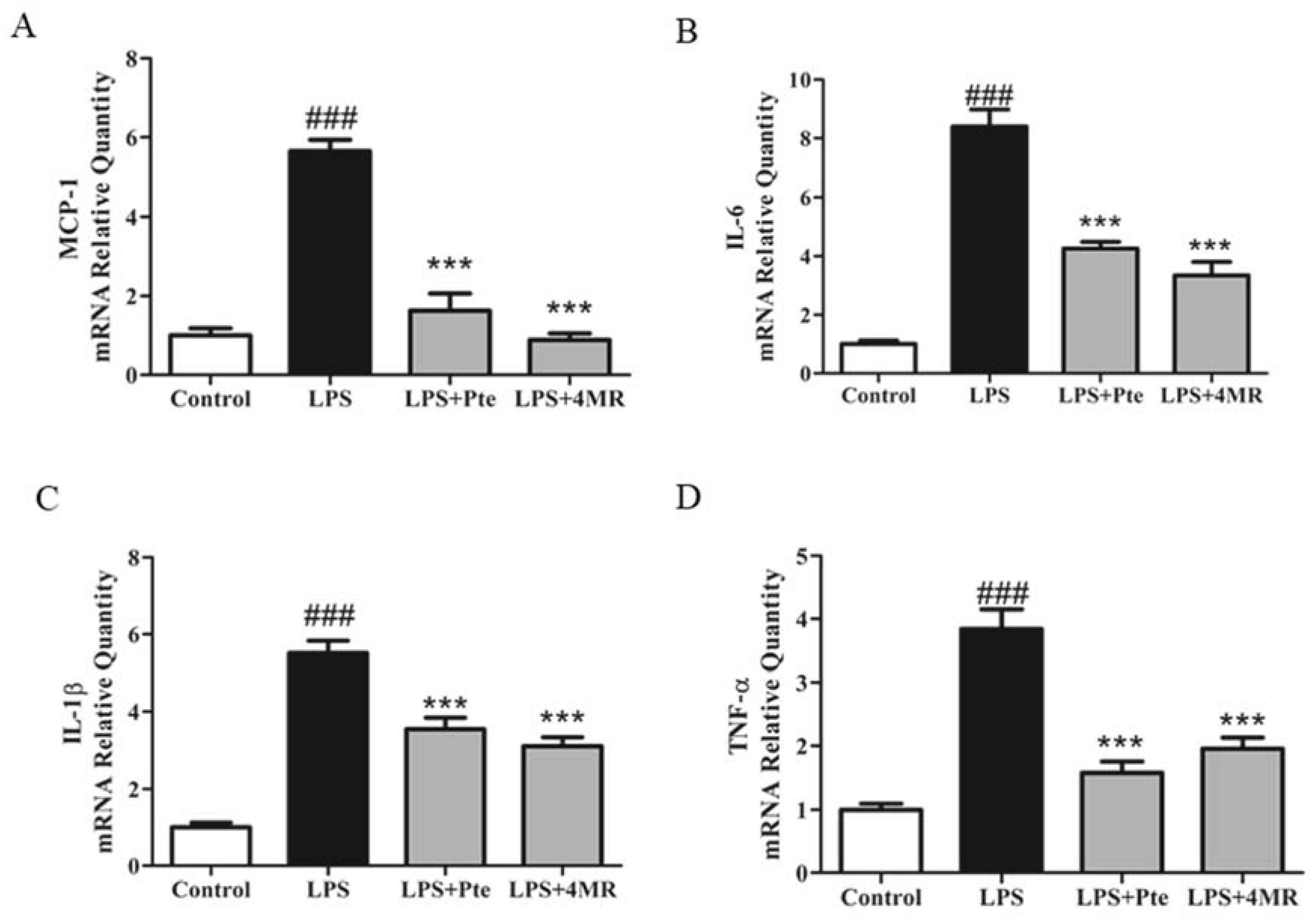
2.3. Pte and 4MR Inhibited LPS-induced Proinflammatory Cytokine Gene Expression
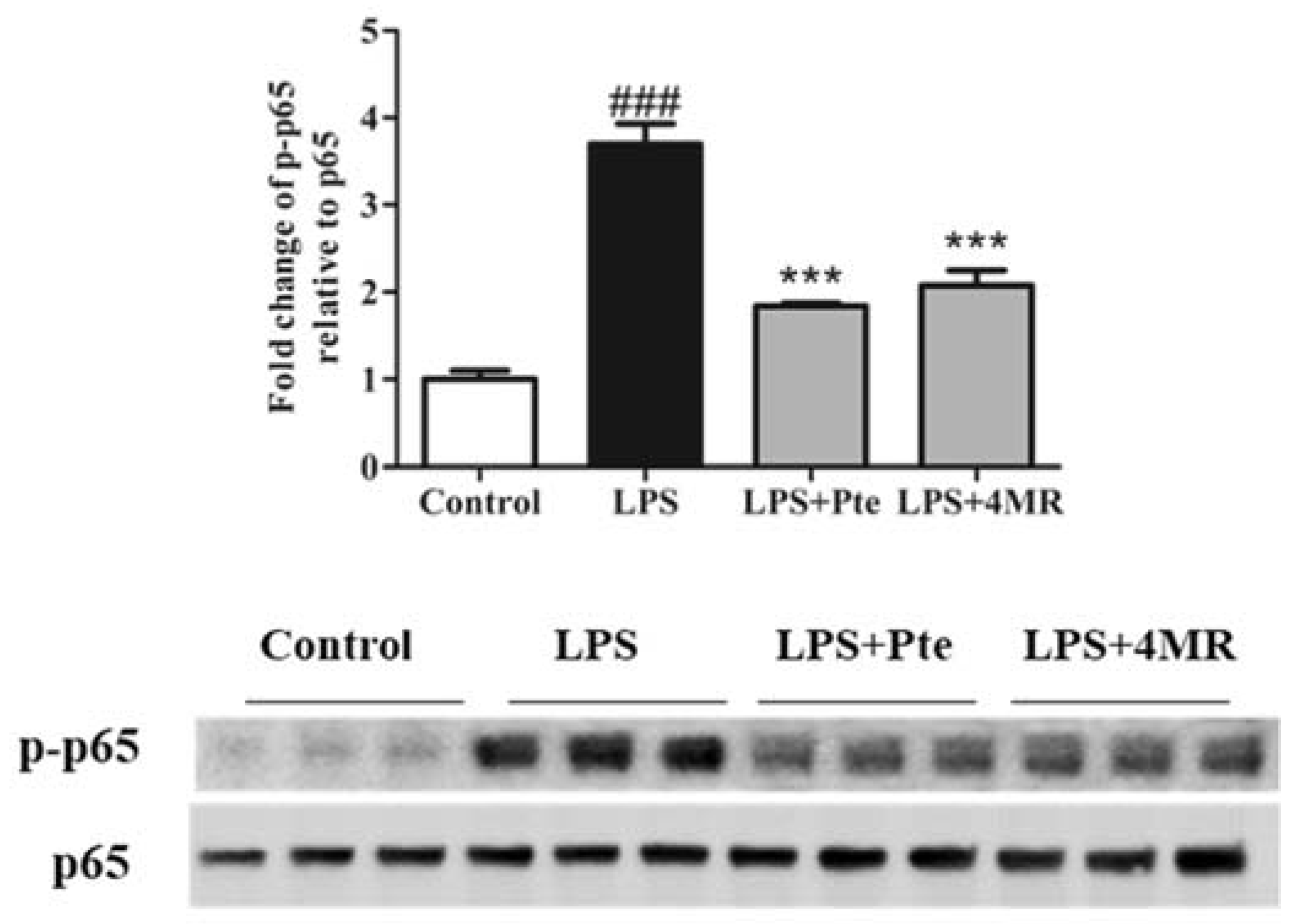
2.4. Pte and 4MR Inhibited LPS-Induced NF-κB Signaling Pathway in RAW 264.7 Macrophages
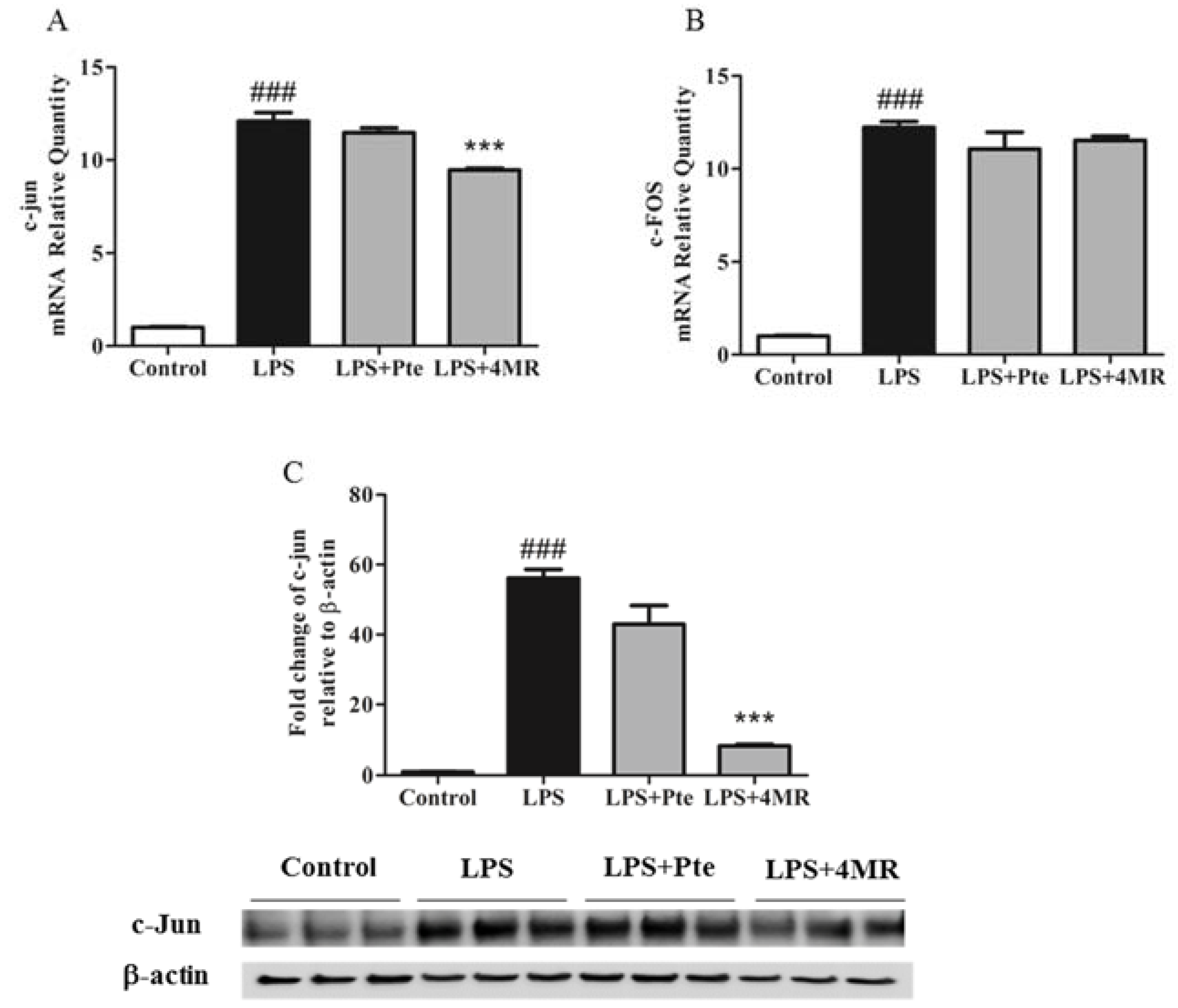
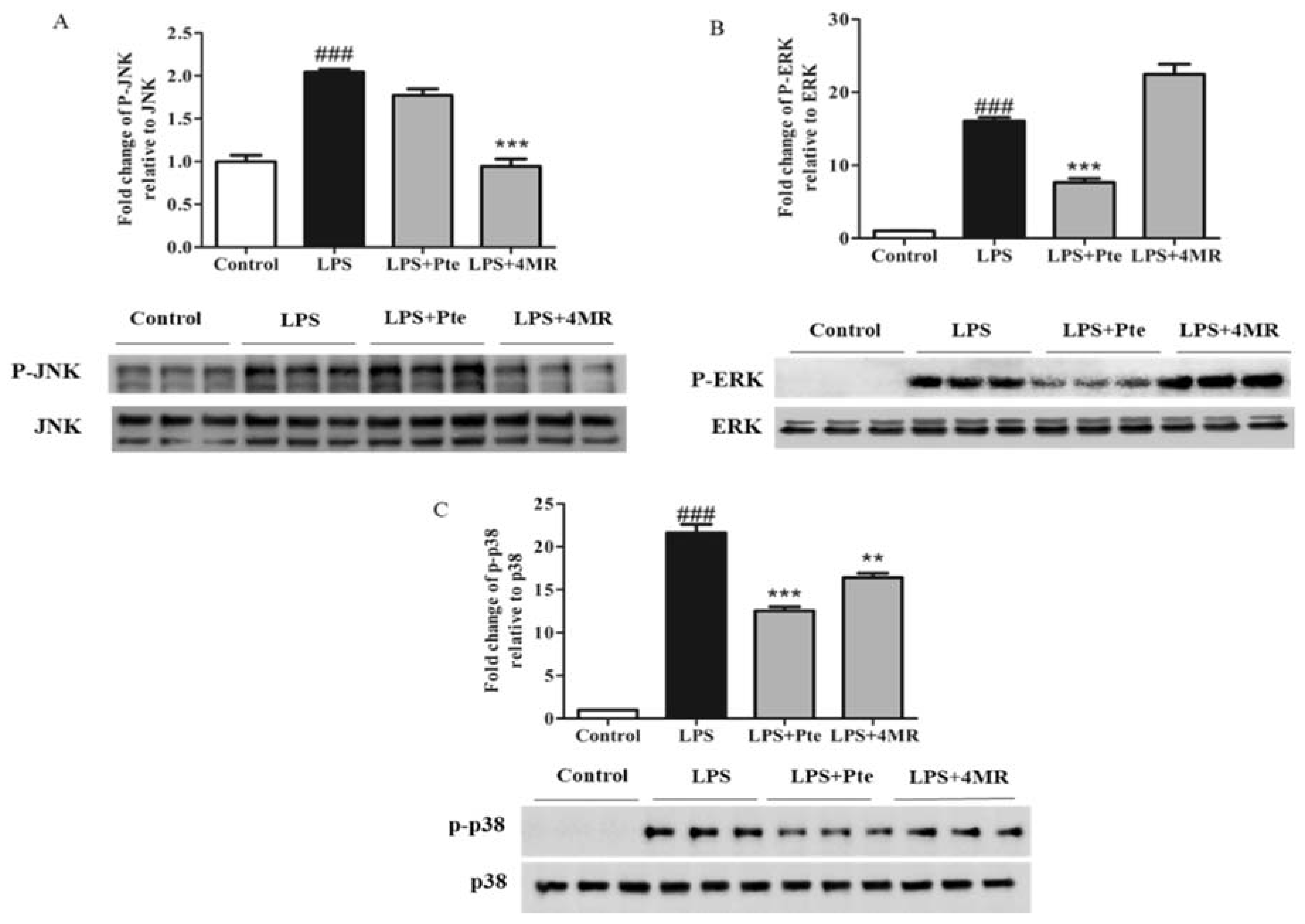
2.5. Pte and 4MR Inhibited LPS-Induced MAPK and AP-1 Pathways in RAW 264.7 Macrophages
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Cell Culture
4.3. Crystal Violet Assay for Cell Viability
4.4. Measurement of NO
4.5. RNA Extraction and Quantitative PCR Analysis
4.6. Western Blot Analysis
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Landskron, G.; De la Fuente, M.; Thuwajit, P.; Thuwajit, C.; Hermoso, M.A. Chronic inflammation and cytokines in the tumor microenvironment. J. Immunol. Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Campisi, J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A-Biol. Sci. Med. Sci. 2014, 69, S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Dehdashtian, E.; Mehrzadi, S.; Yousefi, B.; Hosseinzadeh, A.; Reiter, R.J.; Safa, M.; Ghaznavi, H.; Naseripour, M. Diabetic retinopathy pathogenesis and the ameliorating effects of melatonin; involvement of autophagy, inflammation and oxidative stress. Life Sci. 2018, 193, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, C.; Giacomin, P.; Navarro, S.; Miller, C.; Loukas, A.; Wangchuk, P. A medicinal plant compound, capnoidine, prevents the onset of inflammation in a mouse model of colitis. J. Ethnopharmacol. 2018, 211, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Gravallese, E.M. Mediators of inflammation and bone remodeling in rheumatic disease. Semin. Cell Dev. Biol. 2016, 49, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Chrysohoou, C.; Kollia, N.; Tousoulis, D.C. The link between depression and atherosclerosis through the pathways of inflammation and endothelium dysfunction. Maturitas 2018, 109, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.L.; Yang, H.L.; Shi, Q. Macrophages and bone inflammation. J. Orthop. Transl. 2017, 10, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Guan, H.; Liu, D.; Wu, X.; Fan, M.; Han, J. Flavonoids from sea buckthorn inhibit the lipopolysaccharide-induced inflammatory response in RAW264.7 macrophages through the MAPK and NF-κB pathways. Food Funct. 2017, 8, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Shin, J.S.; Han, H.S.; Lee, H.H.; Park, J.C.; Lee, K.T. Kaempferol 7-O-β-d-glucoside isolated from the leaves of cudrania tricuspidata inhibits LPS-induced expression of pro-inflammatory mediators through inactivation of NF-κB, AP-1, and JAK-STAT in RAW 264.7 macrophages. Chem. Biol. Interact. 2018, 284, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wu, F.; Jin, Z.; Zhai, Z.; Wang, Y.; Tu, B.; Yan, W.; Tang, T. Plumbagin inhibits LPS-induced inflammation through the inactivation of the nuclear factor-kappa B and mitogen activated protein kinase signaling pathways in RAW 264.7 cells. Food Chem. Toxicol. 2014, 64, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Ding, Y.; Wild, C.; Shen, Q.; Zhou, J. Small molecule inhibitors targeting activator protein 1 (AP-1). Med. Chem. 2014, 57, 6930–6948. [Google Scholar] [CrossRef] [PubMed]
- Hannemann, N.; Jordan, J.; Paul, S.; Reid, S.; Baenkler, H.W.; Sonnewald, S.; Bauerle, T.; Vera, J.; Schett, G.; Bozec, A. The AP-1 transcription factor c-Jun promotes arthritis by regulating cyclooxygenase-2 and arginase-1 expression in macrophages. J. Immunol. 2017, 198, 3605–3614. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.W.; Chiou, W.F.; Chao, S.H.; Lee, M.H.; Chenand, C.C.; Tsai, Y.C. Ligustilide prevents LPS-induced iNOS expression in RAW 264.7 macrophages by preventing ROS production and down-regulating the MAPK, NF-kappa Band AP-1 signaling pathways. Int. Immunopharmacol. 2011, 11, 1166–1172. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.K.; Choi, E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta 2010, 1802, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Tak, P.P.; Firestein, G.S. NF-kappa B: a key role in inflammatory diseases. Clin. Investig. 2001, 107, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Deshmane, S.L.; Kremlev, S.; Amini, S.; Sawaya, B.E. Monocyte chemoattractant protein-1 (MCP-1): An overview. J. Interferon Cytokine Res. 2009, 2, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Covarrubias, A.J.; Horng, T. IL-6 strikes a balance in metabolic inflammation. Cell Metab. 2014, 19, 898–899. [Google Scholar] [CrossRef] [PubMed]
- Scholz, C.C.; Cavadas, M.A.; Tambuwala, M.M.; Hams, E.; Rodríguez, J.; Kriegsheim, A.; Cotter, P.; Bruning, U.; Fallon, P.G.; Cheong, A.; et al. Regulation of IL-1β-induced NF-κB by hydroxylases links key hypoxic and inflammatory signaling pathways. Proc. Natl. Acad. Sci. USA 2013, 46, 18490–18495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.L.; Sankar, G. Molecular mechanisms of NF-kappaB activation induced by bacterial lipopolysaccharide through Toll-like receptors. J. Endotoxin Res. 2000, 6, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; DeCastro1, A.J.; Lee1, H.J.; Smolarek1, A.K.; So, J.Y.; Simi, B.; Wang, C.X.; Zhou, R.; Rimando, A.M.; Suh, N. Dietary intake of pterostilbene, a constituent of blueberries, inhibits the β-catenin/p65 downstream signaling pathway and colon carcinogenesis in rats. Carcinogenesis 2010, 31, 1272–1278. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, C.L.; Wang, T.J.; Li, X.F. Synthesis of resveratrol analogs/stilbene derivatives and their nitric oxide inhibitory and radical scavenging activities. Chem. Res. Chin. Univ. 2014, 30, 941–946. [Google Scholar] [CrossRef]
- Pan, M.H.; Chang, Y.H.; Tsai, M.L.; Lai, C.S.; Ho, S.Y.; Badmaev, V.; Ho, C.T. Pterostilbene suppressed lipopolysaccharide-induced up-expression of iNOS and COX-2 in murine macrophages. Agric. Food Chem. 2008, 56, 7502–7509. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Rimando, A.M.; Lee, H.J.; Ji, Y.; Reddy, B.S.; Suh, N. Anti-inflammatory action of pterostilbene is mediated through the p38 mitogen-activated protein kinase pathway in colon cancer cells. Cancer Prev. Res. 2009, 2, 650. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.N.; Al-Omran, A.; Parvathy, S.S. Role of nitric oxide in inflammatory diseases. Inflammopharmacology 2007, 15, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Stark, G.R. NF-kB-dependent signaling pathways. Exp. Hematol. 2002, 30, 285–296. [Google Scholar] [CrossRef]
- Ahn, S.; Singh, P.; Jang, M.; Kim, Y.J.; Castro-Aceituno, V.; Simu, S.Y.; Kim, Y.J.; Yang, D.C. Gold nanoflowers synthesized using Acanthopanacis cortex extract inhibit inflammatory mediators in LPS-induced RAW264.7 macrophages via NF-kB and AP-1 pathways. Colloids Surf. B 2018, 162, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Liu, Z.G.; Zandi, E. AP-1 function and regulation. Curr. Opin. Cell Biol. 1997, 9, 240–246. [Google Scholar] [CrossRef]
- Yang, G.; Ham, I.; Choi, H.Y. Anti-inflammatory effect of prunetin via the suppression of NF-kappa B pathway. Food Chem. Toxicol. 2013, 58, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lenardo, M.; Baltimore, D. 30 years of NF-κB: A blossoming of relevance to human pathobiology. Cell 2017, 168, 37–57. [Google Scholar] [CrossRef] [PubMed]
- Endale, M.; Park, S.C.; Kim, S.; Kim, S.H.; Yang, Y.; Choand, J.Y.; Rhee, M.H. Quercetin disrupts tyrosine-phosphorylated phosphatidylinositol 3-kinase and myeloid differentiation factor-88 association, and inhibits MAPK/AP-1 and IKK/NF-kappa B-induced inflammatory mediators production in RAW 264.7 cells. Immunobiology 2013, 218, 1452–1467. [Google Scholar] [CrossRef] [PubMed]
- Jo, A.R.; Han, H.S.; Seo, S.; Shin, J.S.; Lee, J.Y.; Kim, H.J.; Lee, K.T. Inhibitory effect of moschamine isolated from carthamus tinctorius on LPS-induced inflammatory mediators via AP-1 and STAT1/3 inactivation in RAW 264.7 macrophages. Bioorg. Med. Chem. Lett. 2017, 27, 5245–5251. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.A.; Guan, X.Q.; Reis, J.C.; Papasian, C.J.; Jabre, S.; Morrison, D.C.; Qureshi, N. Inhibition of nitric oxide and inflammatory cytokines in LPS-stimulated murine macrophages by resveratrol, a potent proteasome inhibitor. Lipid Health Dis. 2012, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.S.; Kang, S.I.; Park, D.B.; Kim, S.J. Resveratrol suppresses inflammatory responses and improves glucose uptake in adipocytes interacted with macrophages. Genes Genom. 2016, 38, 137–143. [Google Scholar] [CrossRef]
- Seo, M.J.; Lee, Y.J.; Hwang, J.H.; Kim, K.J.; Lee, B.Y. The inhibitory effects of quercetin on obesity and obesity-induced inflammation by regulation of MAPK signaling. J. Nutr. Biochem. 2015, 26, 1308–1316. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds pterostilbene and 4’-methoxyresveratrol are available from the authors. |


| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| MCP-1 | AGCTCTTTCCTCCACCA | CTACAGCTTCTTTGGGACACCT |
| IL-6 | AGCCAGAGTCCTTCAGAGAGAT | GCACTAGGTTTGCCG AGTAGAT |
| IL-1β | GCAACTGTTCCTGAACTCAACT | ATCTTTTGGGGTCCGTCAACT |
| TNF-α | CACCACGCTCTTCTGTCTACTG | CTTGAGATCCATCGCGTTG |
| iNOS | GGCAGCCTGTGAGACCTTTG | GCATTGGAAGTGAAGCGTTTC |
| c-Jun | CCTTCTACGACGATGCCCTC | AGAAGGTCCGAGTTCTTGGC |
| c-Fos | CGGGTTTCAACGCCGACTA | TGGCACTAGAGACGGACAGAT |
| 18S | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Y.; Liu, K.; Zhao, Y.; Hu, X.; Wang, M. Pterostilbene and 4′-Methoxyresveratrol Inhibited Lipopolysaccharide-Induced Inflammatory Response in RAW264.7 Macrophages. Molecules 2018, 23, 1148. https://doi.org/10.3390/molecules23051148
Yao Y, Liu K, Zhao Y, Hu X, Wang M. Pterostilbene and 4′-Methoxyresveratrol Inhibited Lipopolysaccharide-Induced Inflammatory Response in RAW264.7 Macrophages. Molecules. 2018; 23(5):1148. https://doi.org/10.3390/molecules23051148
Chicago/Turabian StyleYao, Yun, Kehai Liu, Yueliang Zhao, Xiaoqian Hu, and Mingfu Wang. 2018. "Pterostilbene and 4′-Methoxyresveratrol Inhibited Lipopolysaccharide-Induced Inflammatory Response in RAW264.7 Macrophages" Molecules 23, no. 5: 1148. https://doi.org/10.3390/molecules23051148
APA StyleYao, Y., Liu, K., Zhao, Y., Hu, X., & Wang, M. (2018). Pterostilbene and 4′-Methoxyresveratrol Inhibited Lipopolysaccharide-Induced Inflammatory Response in RAW264.7 Macrophages. Molecules, 23(5), 1148. https://doi.org/10.3390/molecules23051148







