Lipase-Catalyzed Synthesis of Sucrose Monolaurate and Its Antibacterial Property and Mode of Action against Four Pathogenic Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Lipase-Catalyzed Transesterification for SML in Ionic Liquid
2.2.1. Synthesis of [3CIM(EO)][NTf2]
2.2.2. General Procedure of the Lipase-Catalyzed Transesterification in the Ionic Liquid
2.2.3. Analytical Procedure
2.3. Bacterial Strains and Growing Conditions
2.4. Antimicrobial Activity
2.4.1. Minimum Inhibitory Concentration (MIC) and Minimum Bactericide Concentration (MBC)
2.4.2. Growth Curve and Time–Kill Kinetics Analysis
2.5. Antibacterial Mechanism
2.5.1. Propidium Iodide Uptake Test
2.5.2. Cell Constituents’ Release
2.5.3. Cytoplasmic β-Galactosidase Released from Cells after SML Treatment
2.5.4. Evaluation of Zeta Potential
2.5.5. Effect of SML on Bacterial DNA
2.5.6. Biofilm Formation Assay
2.5.7. Scanning Electron Microscopy (SEM) Analysis
2.6. Statistical Analysis
3. Results and Discussion
3.1. Lipase-Catalyzed Transesterification in Novel Functionalized Ionic Liquid
3.2. MICs and MBCs of SML
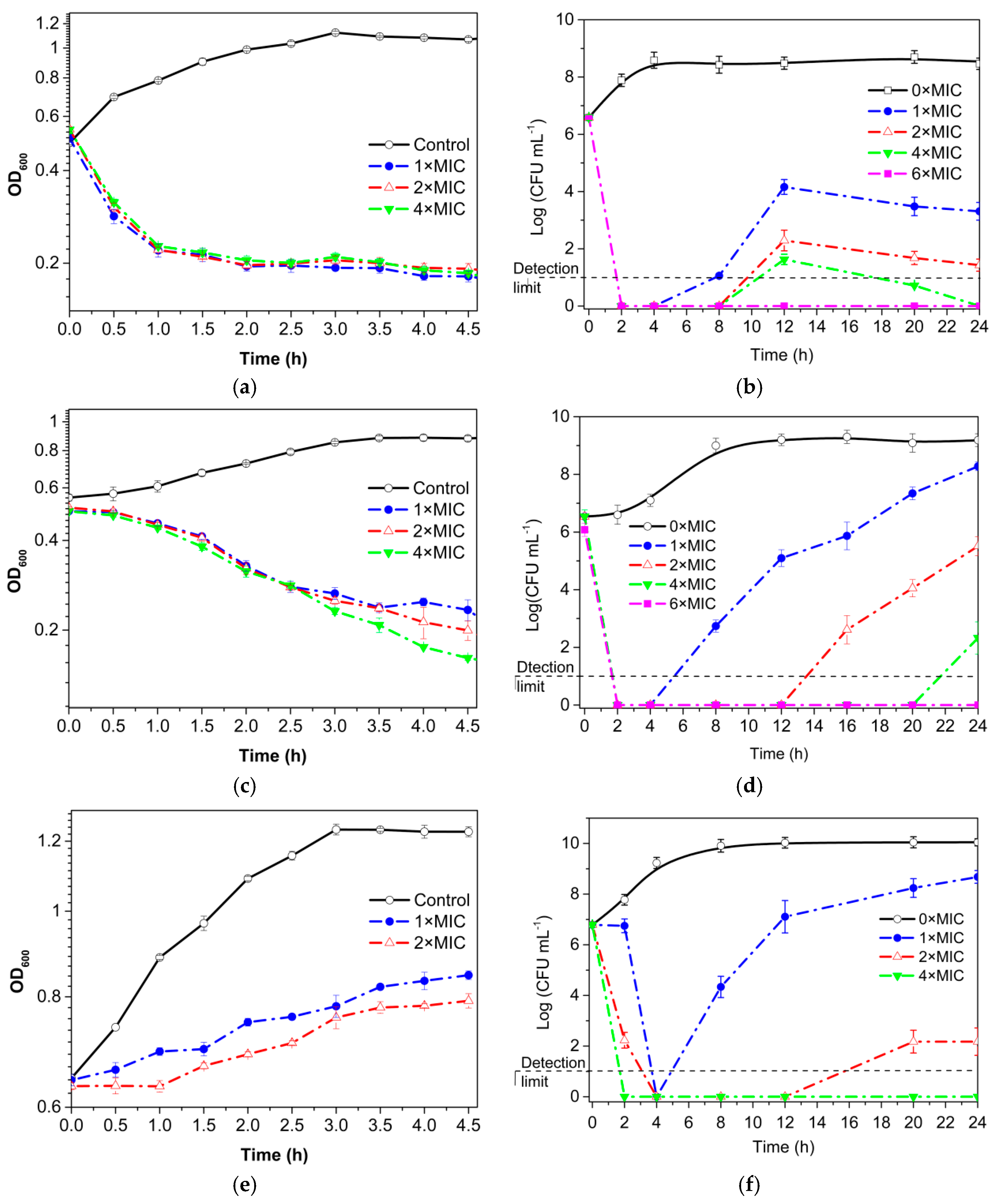
3.3. Growth Curve and Time–Kill Analysis
3.4. Permeability of Cell Membrane
3.5. Scanning Electron Microscopy (SEM) Analysis
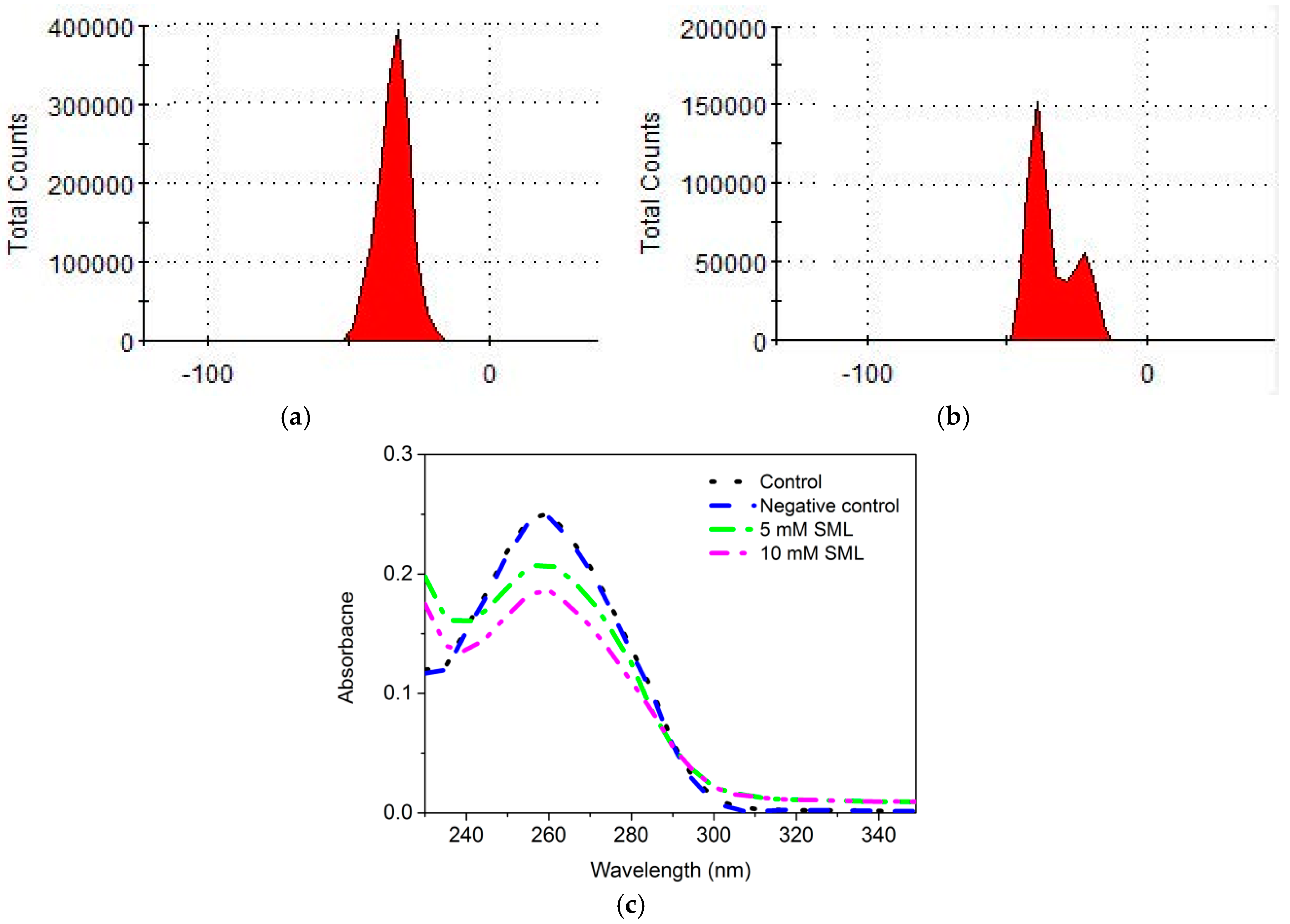
3.6. Effects of SML on the Apparent Zeta Potential of Cells
3.7. Effect of SML on DNA of L. monocytogenes
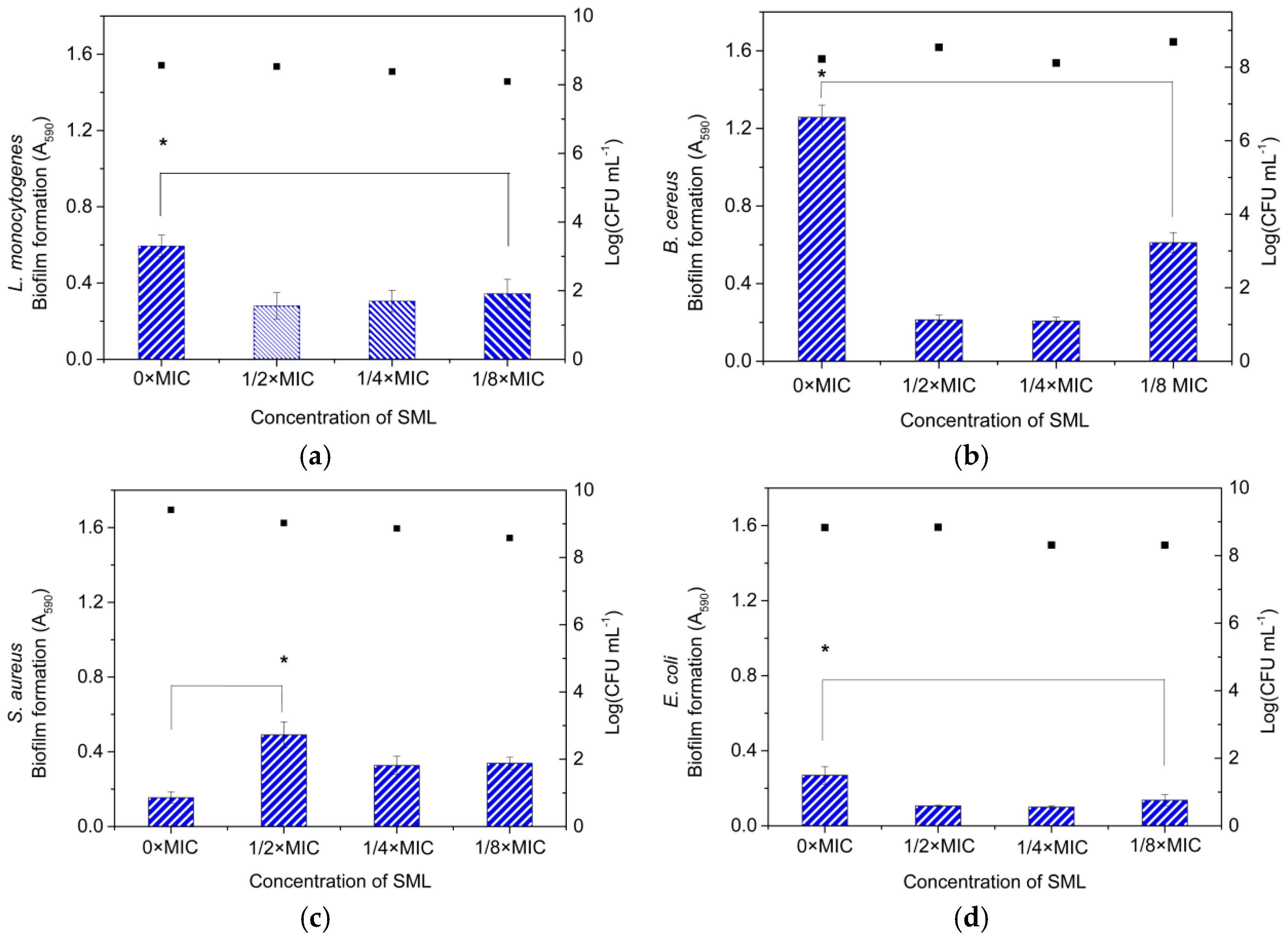
3.8. Effect of SML at Sub-MICs on the Inhibition of Biofilm Formation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rantwijk, F.V.; Lau, R.M.; Sheldon, R.A. Biocatalytic transformations in ionic liquids. Trends Biotechnol. 2003, 21, 131–138. [Google Scholar] [CrossRef]
- Moniruzzaman, M.; Nakashima, K.; Kamiya, N.; Goto, M. Recent advances of enzymatic reactions in ionic liquids. Biochem. Eng. J. 2010, 48, 295–314. [Google Scholar] [CrossRef]
- Quijano, G.; Couvert, A.; Amrane, A. Ionic liquids: Applications and future trends in bioreactor technology. Bioresour. Technol. 2010, 101, 8923–8930. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.G.; Liu, Y.L.; Lai, P.Y.; Tseng, M.C.; Tseng, M.J.; Li, Y.D.; Chu, Y.H. Ionic liquids promote PCR amplification of DNA. Chem. Commun. 2012, 48, 5325–5327. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.G.; Wu, Y.; Lu, X.Y.; Ren, Y.P.; Wang, Q.; Zhu, C.M.; Yu, D.; Wang, H. Lipase-catalyzed esterification of ferulic acid with lauryl alcohol in ionic liquids and antibacterial properties in vitro against three food-related bacteria. Food Chem. 2017, 220, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Monk, J.D.; Beuchat, L.R.; Hathcox, A.K. Inhibitory effects of sucrose monolaurate, alone and in combination with organic acids, on Listeria monocytogenes and Staphylococcus aureus. J. Appl. Bacteriol. 1996, 81, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.M.; Luedecke, L.O.; Swanson, B.G. Inhibition of microorganisms in salad dressing by sucrose and methylglucose fatty acid monoesters. J. Food Process. Preserv. 2003, 27, 285–298. [Google Scholar] [CrossRef]
- Favier, G.I.; Escudero, M.E.; de Guzmán, A.M. Inhibitory effects of sucrose fatty acid esters on survival of Yersinia enterocolitica on eggshell surface and use of blue lake as indicator of bacterial penetration into eggs. J. Appl. Microbiol. 2009, 106, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Arima, K. Inhibitory effect of sucrose ester of lauric acid on the growth of Escherichia coli. Biochem. Biophys. Res. Commun. 1971, 42, 596–601. [Google Scholar] [CrossRef]
- Ferrer, M.; Soliveri, J.; Plou, F.J.; López-Cortes, N.; Reyes-Duarte, D.; Christensen, M.; Copa-Patino, J.L.; Ballesteros, A. Synthesis of sugar esters in solvent mixtures by lipases from Thermomyces lanuginosus and Candida antarctica B, and their antimicrobial properties. Enzym. Microb. Technol. 2005, 36, 391–398. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, H.Y.; Hao, T.Y.; Li, S. In vitro antibacterial activities and mechanism of sugar fatty acid esters against five food-related bacteria. Food Chem. 2015, 187, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Conley, A.J.; Kabara, J.J. Antimicrobial action of esters of polyhydric alcohols. Antimicrob. Agents Chemother. 1973, 4, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Habulin, M.; Šabeder, S.; Knez, Ž. Enzymatic synthesis of sugar fatty acid esters in organic solvent and in supercritical carbon dioxide and their antimicrobial activity. J. Supercrit. Fluids 2008, 45, 338–345. [Google Scholar] [CrossRef]
- Walsh, M.K.; Bombyk, R.A.; Wagh, A.; Bingham, A.; Berreau, L.M. Synthesis of lactose monolaurate as influenced by various lipases and solvents. J. Mol. Catal. B Enzym. 2009, 60, 171–177. [Google Scholar] [CrossRef]
- Wilson, B.; Abraham, G.; Manju, V.S.; Mathew, M.; Vimala, B.; Sundaresan, S.; Nambisan, B. Antimicrobial activity of Curcuma zedoaria and Curcuma malabarica tubers. J. Ethnopharmacol. 2005, 99, 147–151. [Google Scholar] [CrossRef] [PubMed]
- D’Arrigo, M.; Ginestra, G.; Mandalari, G.; Furneri, P.M.; Bisignano, G. Synergism and postantibiotic effect of tobramycin and Melaleuca alternifolia (tea tree) oil against Staphylococcus aureus and Escherichia coli. Phytomedicine 2010, 17, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Park, I.K.; Kang, D.H. Effect of electropermeabilization by ohmic heating for inactivation of Escherichia coli O157:H7, Salmonella enterica serovar Typhimurium, and Listeria monocytogenes in buffered peptone water and apple juice. Appl. Environ. Microb. 2013, 79, 7122–7129. [Google Scholar] [CrossRef] [PubMed]
- Diao, W.R.; Hu, Q.P.; Zhang, H.; Xu, J.G. Chemical composition, antibacterial activity and mechanism of action of essential oil from seeds of fennel (Foeniculum vulgare Mill.). Food Control 2014, 35, 109–116. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Kong, B.H. Study on antibacterial mechanism of clove extract. Food Sci. 2009, 30, 88–91. [Google Scholar]
- Borges, A.; Ferreira, C.; Saavedra, M.J.; Simões, M. Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb. Drug Resist. 2013, 19, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Narisawa, N.; Furukawa, S.; Ogihara, H.; Yamasaki, M. Estimation of the biofilm formation of Escherichia coli K-12 by the cell number. J. Biosci. Bioeng. 2005, 99, 78–80. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.H.; Zheng, J.; Zhang, X.L.; Hu, Q.D.; Qian, R.J. Salicylic acid alleviates the adverse effects of salt stress on Dianthus superbus (Caryophyllaceae) by activating photosynthesis, protecting morphological structure, and enhancing the antioxidant system. Front. Plant Sci. 2017, 8, 600. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Lalonde, J.J.; Momongan, M.; Bergbreiter, D.E.; Wong, C.H. Lipase-catalyzed irreversible transesterifications using enol esters as acylating reagents: Preparative enantio- and regioselective syntheses of alcohols, glycerol derivatives, sugars and organometallics. J. Am. Chem. Soc. 1988, 110, 7200–7205. [Google Scholar] [CrossRef]
- Ferrer, M.; Cruces, M.A.; Bernabé, M.; Ballesteros, A.; Plou, F.J. Lipase-Catalyzed Regioselective Acylation of Sucrose in Two-Solvent Mixtures. Biotechnol. Bioeng. 1999, 65, 10–16. [Google Scholar] [CrossRef]
- Zhao, H.; Song, Z.; Olubajo, O. High transesterification activities of immobilized proteases in new ether-functionalized ionic liquids. Biotechnol. Lett. 2010, 32, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Potier, P.; Bouchu, A.; Gagnaire, J.; Queneau, Y. Proteinase N-catalysed regioselective esterification of sucrose and other mono- and disaccharides. Tetrahedron Asymmetry 2001, 12, 2409–2419. [Google Scholar] [CrossRef]
- Wagh, A.; Shen, S.; Shen, F.A.; Miller, C.D.; Walsh, M.K. Effect of lactose monolaurate on pathogenic and nonpathogenic bacteria. Appl. Environ. Microbiol. 2012, 78, 3465–3468. [Google Scholar] [CrossRef] [PubMed]
- Jouki, M.; Mortazavi, S.A.; Yazdi, F.T.; Koocheki, A. Optimization of extraction, antioxidant activity and functional properties of quince seed mucilage by RSM. Int. J. Biol. Macromol. 2014, 66, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Tsuchido, T.; Ahn, Y.H.; Takano, M. Lysis of bacillus subtilis cells by glycerol and sucrose esters of fatty acids. Appl. Environ. Microbiol. 1987, 53, 505–508. [Google Scholar] [PubMed]
- Ferro, B.E.; Van, I.J.; Wattenberg, M.; Van, S.D.; Mouton, J.W. Time–kill kinetics of antibiotics active against rapidly growing mycobacteria. J. Antimicrob. Chemother. 2015, 70, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Osawa, M.; Erickson, H.P. Turgor pressure and possible constriction mechanisms in bacterial division. Front. Microbiol. 2018, 9, 111. [Google Scholar] [CrossRef] [PubMed]
- Devulapalle, K.S.; de Segura, A.G.; Ferrer, M.; Alcalde, M.; Mooser, G.; Plou, F.J. Effect of carbohydrate fatty acid esters on Streptococcus sobrinus and glucosyltransferase activity. Carbohydr. Res. 2004, 339, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Becerra, N.; Nuez, L.R.D.; Zanocco, A.L.; Lemp, E.; Günther, G. Solubilization of dodac small unilamellar vesicles by sucrose esters: A fluorescence study. Colloids Surf. A Physicochem. Eng. Asp. 2006, 272, 2–7. [Google Scholar] [CrossRef]
- Palmer, J.; Flint, S.; Brooks, J. Bacterial cell attachment, the beginning of a biofilm. J. Ind. Microbiol. Biotechnol. 2007, 34, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Ahimou, F.; Denis, F.A.; Touhami, A.A.; Dufrêne, Y.F. Probing microbial cell surface charges by atomic force microscopy. Langmuir 2002, 18, 9937–9941. [Google Scholar] [CrossRef]
- Girard, L.P.; Ceri, H.; Olson, A.P.M.; Sepandj, F. MIC versus MBEC to determine the antibiotic sensitivity of Staphylococcus aureus in peritoneal dialysis peritonitis. Perit. Dial. Int. 2010, 30, 652–656. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
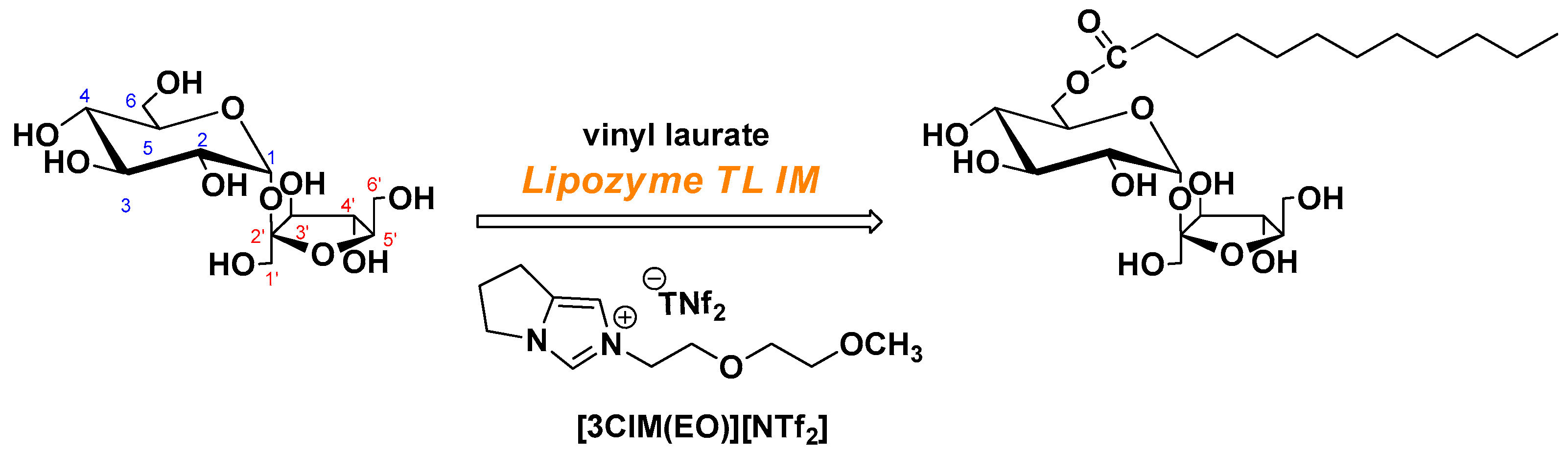



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, S.-Y.; Shi, Y.-G.; Wu, Y.; Bian, L.-Q.; Zhu, Y.-J.; Huang, X.-Y.; Pan, Y.; Zeng, L.-Y.; Zhang, R.-R. Lipase-Catalyzed Synthesis of Sucrose Monolaurate and Its Antibacterial Property and Mode of Action against Four Pathogenic Bacteria. Molecules 2018, 23, 1118. https://doi.org/10.3390/molecules23051118
Shao S-Y, Shi Y-G, Wu Y, Bian L-Q, Zhu Y-J, Huang X-Y, Pan Y, Zeng L-Y, Zhang R-R. Lipase-Catalyzed Synthesis of Sucrose Monolaurate and Its Antibacterial Property and Mode of Action against Four Pathogenic Bacteria. Molecules. 2018; 23(5):1118. https://doi.org/10.3390/molecules23051118
Chicago/Turabian StyleShao, Shi-Yin, Yu-Gang Shi, Yu Wu, Li-Qing Bian, Yun-Jie Zhu, Xin-Ying Huang, Ying Pan, Lu-Yao Zeng, and Run-Run Zhang. 2018. "Lipase-Catalyzed Synthesis of Sucrose Monolaurate and Its Antibacterial Property and Mode of Action against Four Pathogenic Bacteria" Molecules 23, no. 5: 1118. https://doi.org/10.3390/molecules23051118
APA StyleShao, S.-Y., Shi, Y.-G., Wu, Y., Bian, L.-Q., Zhu, Y.-J., Huang, X.-Y., Pan, Y., Zeng, L.-Y., & Zhang, R.-R. (2018). Lipase-Catalyzed Synthesis of Sucrose Monolaurate and Its Antibacterial Property and Mode of Action against Four Pathogenic Bacteria. Molecules, 23(5), 1118. https://doi.org/10.3390/molecules23051118



