Prognostic Significance of Activated Leukocyte Cell Adhesion Molecule (ALCAM) in Association with Promoter Methylation of the ALCAM Gene in Breast Cancer
Abstract
1. Introduction
2. Results
2.1. Clinicopathologic Characteristics
2.2. Association of Methylation Status of the ALCAM Gene and ALCAM Expression
2.3. Association of Methylation Levels of the ALCAM Gene and ALCAM Expression with Inflammatory Markers in Tumor Tissues
2.4. Association of Methylation Levels of the ALCAM Gene and Its Expression with the Clinicopathologic Characteristics
2.5. Association between Inflammatory Markers and the Clinicopathologic Characteristics in Breast Cancer
2.6. Discussion
3. Materials and Methods
3.1. Patients and Materials
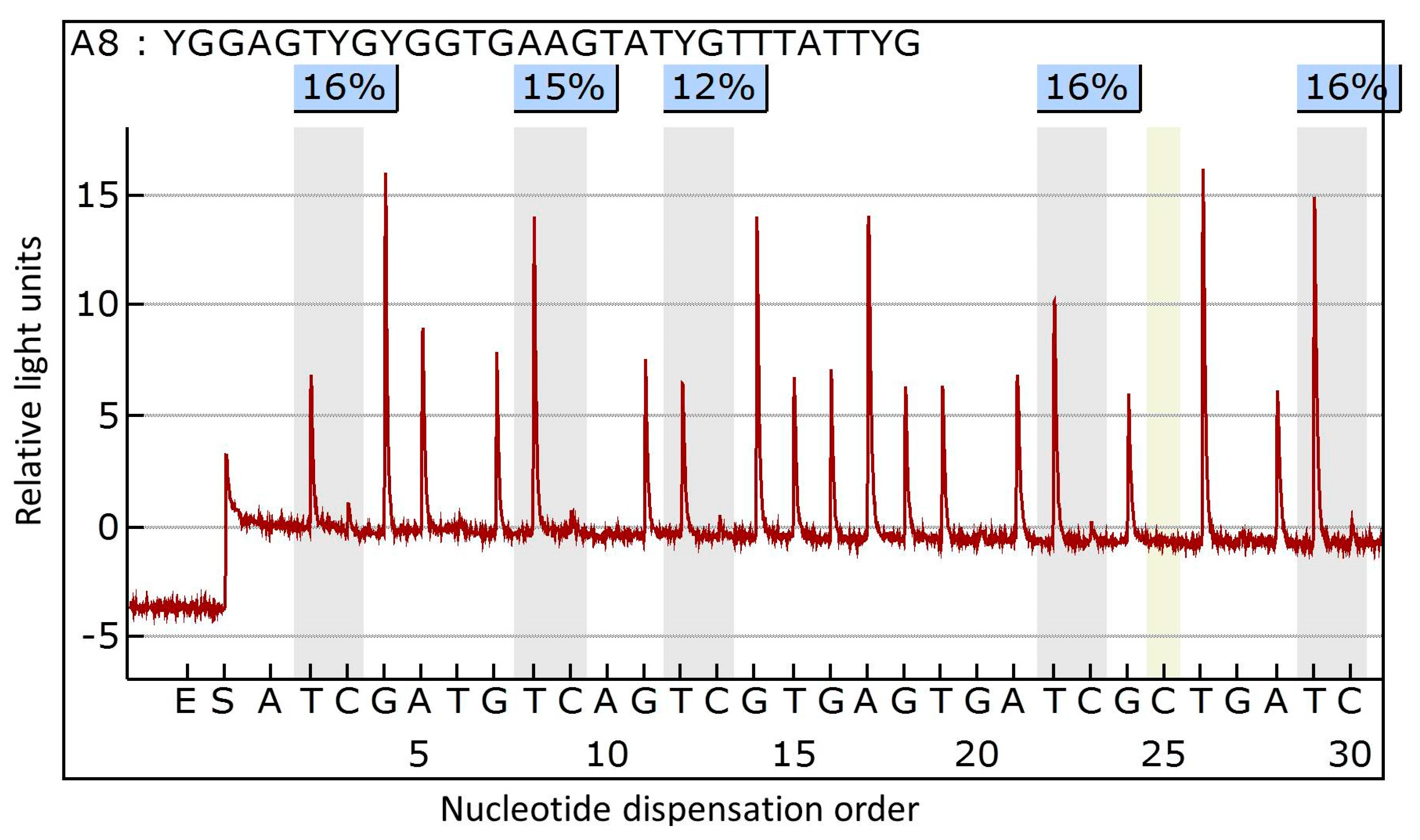
3.2. DNA Extraction and Methylation Analysis
3.3. RNA Extraction and Analysis of ALCAM Transcripts
3.4. Tissue Microarray and Immunohistochemistry
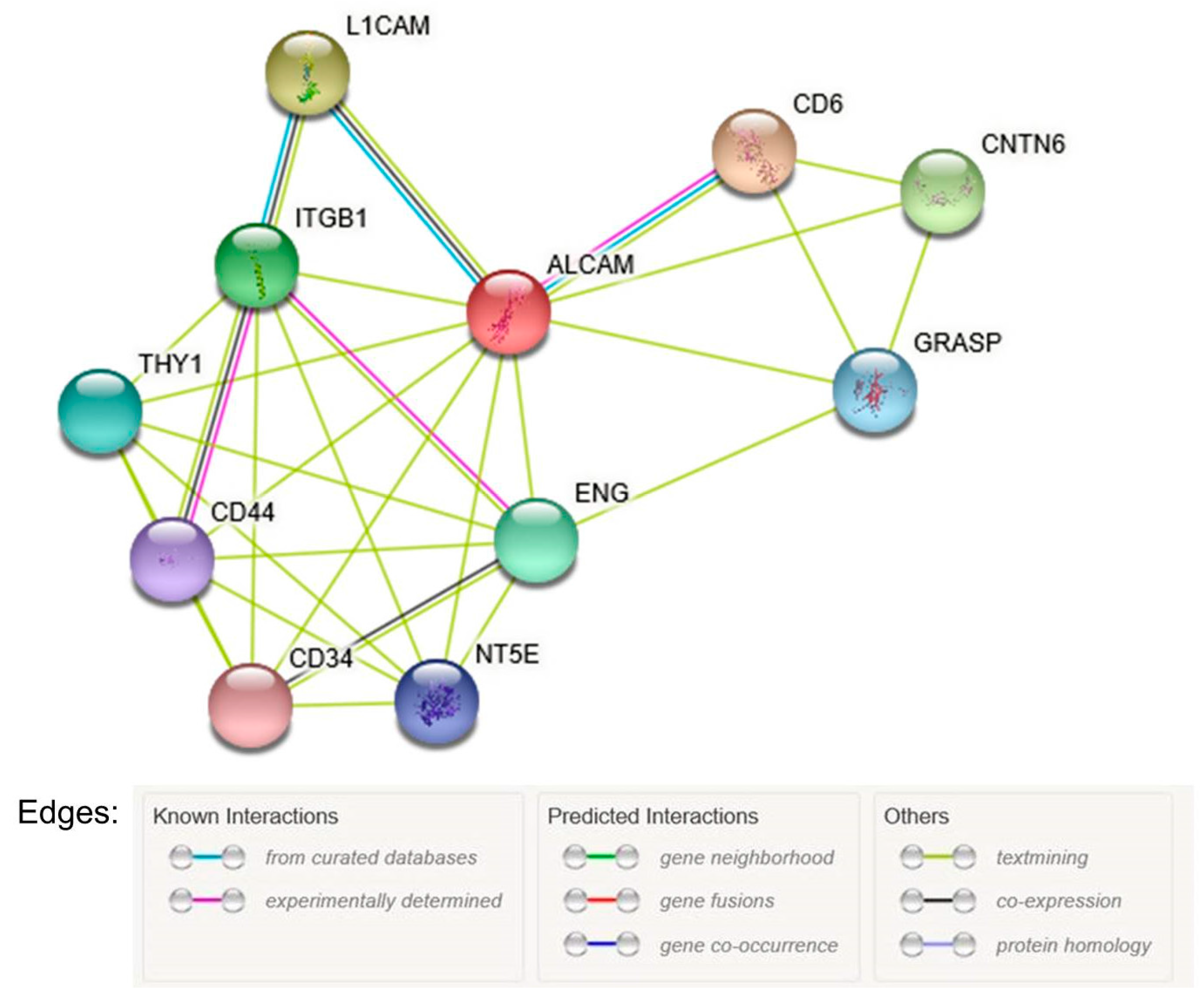
3.5. Bioinformatic Analysis
3.6. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Makrilia, N.; Kollias, A.; Manolopoulos, L.; Syrigos, K. Cell adhesion molecules: Role and clinical significance in cancer. Cancer Investig. 2009, 27, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Okegawa, T.; Pong, R.C.; Li, Y.; Hsieh, J.T. The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochim. Pol. 2004, 51, 445–457. [Google Scholar] [PubMed]
- Cohen, M.B.; Grieblin, T.L.; Ahaghotu, C.A.; Rokhlin, O.W.; ROSS, J.S. Cellular adhesion molecules in urologic malignancies. Am. J. Clin. Pathol. 1997, 107, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Epenetos, A.A.; Pigantelli, M. Cell Adhesion Molecules in Cancer and Inflammation; Hardwood Academic Publishers: Chur, Switzerland, 1995; p. 230. [Google Scholar]
- Bowen, M.A.; Patel, D.D.; Li, X.; Modrell, B.; Malacko, A.R.; Wang, W.C.; Marquardt, H.; Neubauer, M.; Pesando, J.M.; Francke, U.; et al. Cloning, mapping, and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J. Exp. Med. 1995, 181, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Van Kempen, L.C.; Nelissen, J.M.; Degen, W.G.; Torensma, R.; Weidle, U.H.; Bloemers, H.P.; Figdor, C.G.; Swart, G.W. Molecular basis for the homophilic activated leukocyte cell adhesion molecule (ALCAM)-ALCAM interaction. J. Biol. Chem. 2001, 276, 25783–25790. [Google Scholar] [CrossRef] [PubMed]
- Ohneda, O.; Ohneda, K.; Arai, F.; Lee, J.; Miyamoto, T.; Fukushima, Y.; Dowbenko, D.; Lasky, L.A.; Suda, T. ALCAM (CD66): Its role in hematopoietic and endothelial development. Blood 2001, 98, 2134–2142. [Google Scholar] [CrossRef] [PubMed]
- Bloemers, H.P.; Swart, G.W. Activated leukocyte cell adhesion molecule/CD166, a marker of tumor progression in primary malignant melanoma of the skin. Am. J. Pathol. 2000, 156, 769–774. [Google Scholar]
- Ishiguro, F.; Murakami, H.; Mizuno, T.; Fujii, M.; Kondo, Y.; Usami, N.; Taniguchi, T.; Yokoi, K.; Osada, H.; Sekido, Y. Membranous expression of activated leukocyte cell adhesion molecule contributes to poor prognosis and malignant phenotypes of non–small-cell lung cancer. J. Surg. Res. 2013, 179, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Shukla, N.K.; Deo, S.V.; Gupta, S.D.; Ralhan, R. MEMD/ALCAM: A potential marker for tumor invasion and nodal metastasis in esophageal squamous cell carcinoma. Oncology 2005, 68, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Knosel, T.; Bellach, J.; Dietel, M.; Kristiansen, G. ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J. Clin. Pathol. 2004, 57, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Weber, H.; Mogler, C.; Bergmann, F.; Schirmacher, P.; Kenngott, H.G.; Matterne, U.; Mollberg, N.; Rahbari, N.N.; Hinz, U.; et al. Increased expression of ALCAM/CD166 in pancreatic cancer is an independent prognostic marker for poor survival and early tumour relapse. Br. J. Cancer 2009, 101, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, G.; Pilarsky, C.; Wissmann, C.; Stephan, C.; Weissbach, L.; Loy, V.; Loening, S.; Dietel, M.; Rosenthal, A. ALCAM/CD166 is up-regulated in low grade prostate cancer and progressively lost in high-grade lesions. Prostate 2003, 54, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; van Bokhoven, A.; Jansen, C.F.J.; Kiemeney, L.A.; Karthaus, H.F.M.; Vriesema, J.; Bussemakers, M.J.G.; Witjes, J.A.; Schalken, J.A. Activated leukocyte cell adhesion molecule (ALCAM) expression is associated with a poor prognosis for bladder cancer patients. UroOncology 2003, 3, 121–129. [Google Scholar] [CrossRef]
- Mezzanzanica, D.; Fabbi, M.; Bagnoli, M.; Staurengo, S.; Losa, M.; Balladore, E.; Alberti, P.; Lusa, L.; Ditto, A.; Ferrini, S.; et al. Subcellular localization of activated leukocyte cell adhesion molecule is a molecular predictor of survival in ovarian carcinoma patients. Clin. Cancer Res. 2008, 14, 1726–1733. [Google Scholar] [CrossRef] [PubMed]
- King, J.A.; Ofori-Acquah, S.F.; Stevens, T.; Al-Mehdi, A.B.; Fodstad, O.; Jiang, W.G. Activated leukocyte cell adhesion molecule in breast cancer: Prognostic indicator. Breast Cancer Res. 2004, 6, R478–R487. [Google Scholar] [CrossRef] [PubMed]
- Burandt, E.; Bari, N.T.; Lebeau, A.; Minner, S.; Burdelski, C.; Jänicke, F.; Müller, V.; Terracciano, L.; Simon, R.; Sauter, G.; et al. Loss of ALCAM expression is linked to adverse phenotype and poor prognosis in breast cancer: A TMA-based immunohistochemical study on 2,197 breast cancer patients. Oncol. Rep. 2014, 32, 2628–2634. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.R.; Dent, C.; Watkins, G.; King, J.A.; Mokbel, K.; Jiang, W.G. Expression of the cell to cell adhesion molecule, ALCAM, in breast cancer patients and the potential link with skeletal metastasis. Oncol. Rep. 2008, 19, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.R.; Jiang, W.G. ALCAM, activated leukocyte cell adhesion molecule, influences the aggressive nature of breast cancer cells, a potential connection to bone metastasis. Anticancer Res. 2010, 30, 1163–1168. [Google Scholar] [PubMed]
- Burkhardt, M.; Mayordomo, E.; Winzer, K.J.; Fritzsche, F.; Gansukh, T.; Pahl, S.; Weichert, W.; Denkert, C.; Guski, H.; Dietel, M.; et al. Cytoplasmic overexpression of ALCAM is prognostic of disease progression in breast cancer. J. Clin. Pathol. 2006, 59, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Hein, S.; Muller, V.; Kohler, N.; Wikman, H.; Krenkel, S.; Streichert, T.; Schweizer, M.; Riethdorf, S.; Assmann, V.; Ihnen, M.; et al. Biologic role of activated leukocyte cell adhesion molecule overexpression in breast cancer cell lines and clinical tumor tissue. Breast Cancer Res. Treat. 2014, 129, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Piao, D.; Jiang, T.; Liu, G.; Wang, B.; Xu, J.; Zhu, A. Clinical implications of activated leukocyte cell adhesion molecule expression in breast cancer. Mol. Biol. Rep. 2012, 39, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.M.; Ferragut, F.; Cárdenas, D.V.M.; Bracalente, C.; Bravo, A.I.; Cagnoni, A.J.; Nuñez, M.; Morosi, L.G.; Quinta, H.R.; Espelt, M.V.; et al. Glycosylation-dependent binding of galectin-8 to activated leukocyte cell adhesion molecule (ALCAM/CD166) promotes its surface segregation on breast cancer cells. Biochim. Biophys. Acta 2016, 1860, 2255–2268. [Google Scholar] [CrossRef] [PubMed]
- King, J.A.; Tan, F.; Mbeunkui, F.; Chambers, Z.; Cantrell, S.; Chen, H.; Alvarez, D.; Shevde, L.A.; Ofori-Acquach, S.F. Mechanisms of transcriptional regulation and prognostic significance of activated leukocyte cell adhesion molecule in cancer. Mol. Cancer 2010, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.J.; Panel Members. Strategies for subtypes—Dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.J.; Simmonds, S.J.; Clarkson, N.G.; Hanrahan, S.; Puklavec, M.J.; Bomb, M.; Barclay, A.N.; Brown, M.H. CD6 regulates T-cell response through activation-dependent recruitment of the positive regulator SLP-76. Mol. Cell. Biol. 2006, 26, 6727–6738. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Widschwendter, M.; Jones, P.A. DNA methylation and breast carcinogenesis. Oncogene 2002, 21, 5462–5482. [Google Scholar] [CrossRef] [PubMed]
- Vairapandi, M. Characterization of DNA demethylation in normal and cancerous cell lines and the regulatory role of cell cycle proteins in human DNA demethylase activity. J. Cell. Biochem. 2004, 91, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Jun, Y.; Kim, J.; Park, H.; Choi, K.S.; Zhang, H.; Park, J.A.; Kwon, J.Y.; Kim, Y.M.; Lee, S.; et al. Integrative analysis of DNA methylation and mRNA expression during differentiation of umbilical cord blood derived mononuclear cells to endothelial cells. Gene 2017, 635, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Baysan, M.; Woolard, K.; Bozdag, S.; Riddick, G.; Kotliarova, S.; Cam, M.C.; Belova, G.I.; Ahs, S.; Zhang, W.; Song, H.; et al. Micro-environment causes reversible changes in DNA methylation and mRNA expression profiles in patient-derived glioma stem cells. PLoS ONE 2014, 9, e94045. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K.; et al. Role of tumor microenvironment in tumorigenesis. J. Cancer 2017, 8, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, A.W.; Joosten, B.; Torensma, R.; Parnes, J.R.; van Leeuwen, F.N.; Figdor, C.G. Long-term engagement of CD6 and ACLAM in essential for T-cell proliferation induced by dendritic cells. Blood 2006, 107, 3212–3220. [Google Scholar] [CrossRef] [PubMed]
- Kudo-Saito, C.; Fuwa, T.; Kawakami, Y. Targeting ALCAM in the cryo-treated tumour microenvironment successfully induces systemic anti-tumour immunity. Eur. J. Cancer 2016, 62, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Obeid, E.; Nanda, R.; Fu, Y.X.; Olopade, O.I. The role of tumor-associated macrophages in breast cancer progression. Int. J. Oncol. 2013, 43, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Dedeurwaerder, S.; Desmedt, C.; Calonne, E.; Singhal, S.K.; Haibe-Kains, B.; Defrance, M.; Michiels, S.; Volkmar, M.; Deplus, R.; Luciani, J.; et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med. 2011, 3, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, S.M.A.; Paish, E.C.; Powe, D.G.; Macmillan, R.D.; Grainge, M.J.; Lee, A.H.S.; Ellis, I.O.; Green, A.R. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 2011, 29, 1949–1955. [Google Scholar] [CrossRef] [PubMed]
- Stanton, S.E.; Adams, S.; Disis, M.L. Variation in the incidence and magnitude of tumor-infiltrating lymphocytes in breast cancer subtypes. JAMA Oncol. 2016, 2, 1354–1360. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Muller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 2010, 28, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Stanton, S.E.; Disis, M.L. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J. Immiother. Cancer 2016, 4, 59. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Shapiro, D.J. The immune system and inflammation in breast cancer. Mol. Cell. Endocrinol. 2014, 382, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.J.; Jeong, H.Y.; Bong, J.G.; Park, S.H.; Oh, H.K. Low methylation levels of the SFRP1 gene are associated with the basal-like subtype of breast cancer. Oncol. Rep. 2013, 29, 1946–1954. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
| ALCAM Methylation Levels | |||||
|---|---|---|---|---|---|
| Tumor | Normal | ||||
| Mean (%) | p-Value | Mean (%) | p-Value | ||
| ALCAM transcript | Negative | 5.17 ± 6.83 | 0.027 | 2.07 ± 0.52 | 0.951 |
| Positive | 2.66 ± 3.34 | 2.04 ± 0.76 | |||
| Inflammatory Markers | ALCAM Methylation | ALCAM Transcripts | |||
|---|---|---|---|---|---|
| Mean Levels (%) | p-Value | Positive Expression, n (%) | p-Value | ||
| TNF-α | Negative | 3.11 ± 3.41 | 0.190 | 6 (37.5) | <0.001 |
| Positive | 3.50 ± 5.30 | 24 (77.4) | |||
| NF-κB p50 | Negative | 9.27 ± 10.51 | 0.318 | 3 (16.7) | <0.001 |
| Positive | 3.12 ± 4.31 | 10 (76.9) | |||
| IL-4 | Negative | 4.20 ± 5.74 | 0.234 | 5 (41.7) | 0.012 |
| Positive | 2.51 ± 3.04 | 19 (90.5) | |||
| Intratumoral inflammation | Negative | 5.23 ± 8.87 | 0.429 | 7 (87.5) | 0.041 |
| Positive | 3.47 ± 4.22 | 17 (60.7) | |||
| Clinicopathologic Characteristics | ALCAM IHC Results | ||||
|---|---|---|---|---|---|
| Negative (%) | Weak (%) | Moderate (%) | Strong (%) | p-Value | |
| Histologic grade | 0.015 | ||||
| 1 | 0.0 | 33.3 | 16.7 | 50.0 | |
| 2 | 9.1 | 27.3 | 36.4 | 27.3 | |
| 3 | 29.4 | 35.3 | 29.4 | 5.9 | |
| HER2 overexpression | 0.021 | ||||
| Negative | 6.7 | 20.0 | 40.0 | 33.3 | |
| Positive | 20.0 | 46.7 | 26.7 | 6.7 | |
| Molecular Subtype | 0.001 | ||||
| Luminal A | 0.0 | 0.0 | 16.7 | 83.3 | |
| Luminal B | 22.2 | 38.9 | 38.9 | 0.0 | |
| HER2 | 0.0 | 50.0 | 25.0 | 25.0 | |
| Basal-like | 0.0 | 50.0 | 50.0 | 0.0 | |
| Clinicopathologic Characteristics | TNFα Expression | ||||
|---|---|---|---|---|---|
| Negative (%) | Weak Positive (%) | Strong Positive (%) | p-Value | ||
| Histologic grade | 1 | 25.0 | 12.5 | 62.5 | 0.039 |
| 2 | 33.3 | 50.0 | 16.7 | ||
| 3 | 37.0 | 18.5 | 44.4 | ||
| Molecular subtype | Luminal A | 33.3 | 33.3 | 33.3 | 0.011 |
| Luminal B | 26.1 | 30.47 | 43.5 | ||
| HER2 | 83.3 | 0.0 | 16.7 | ||
| Basal-like | 33.3 | 0.0 | 66.7 | ||
| HER2 overexpression | Negative | 30.4 | 17.4 | 52.2 | 0.037 |
| Positive | 44.4 | 33.3 | 22.2 | ||
| Bcl 2 expression | Negative | 50.0 | 0.0 | 50. | 0.041 |
| Positive | 30.8 | 30.8 | 38.5 | ||
| Clinicopathologic Characteristics | NF-κB Expression | ||||
|---|---|---|---|---|---|
| Negative (%) | Weak Positive (%) | Strong Positive (%) | p-Value | ||
| ER | Negative | 18.2 | 54.5 | 27.3 | 0.046 |
| Positive | 12.0 | 28.0 | 60.0 | ||
| Molecular subtype | Luminal A | 25.0 | 37.5 | 37.5 | 0.022 |
| Luminal B | 11.8 | 17.6 | 70.6 | ||
| HER2 | 25.0 | 75.0 | 0.0 | ||
| Basal-like | 0.0 | 50.0 | 50.0 | ||
| Clinicopathologic Characteristics | IL-4 Expression | ||||
|---|---|---|---|---|---|
| Negative (%) | Weak Positive (%) | Strong Positive (%) | p-Value | ||
| Stage | I | 50.0 | 0.0 | 50.0 | 0.040 |
| II | 31.3 | 31.3 | 37.5 | ||
| III | 33.3 | 33.3 | 33.3 | ||
| IV | 0.0 | 50.0 | 50.0 | ||
| Tumor size (mean, cm) | 1.83 ± 1.04 | 3.04 ± 0.94 | 1.74 ± 1.10 | 0.029 | |
| Lymphovascular invasion | Negative | 45.0 | 5.0 | 50.0 | 0.003 |
| Positive | 23.1 | 46.2 | 30.8 | ||
| Lymph node metastasis | Negative | 42.9 | 4.8 | 52.4 | 0.002 |
| Positive | 25.0 | 50.0 | 25.0 | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, Y.J.; Oh, H.K.; Park, S.H.; Bong, J.G. Prognostic Significance of Activated Leukocyte Cell Adhesion Molecule (ALCAM) in Association with Promoter Methylation of the ALCAM Gene in Breast Cancer. Molecules 2018, 23, 131. https://doi.org/10.3390/molecules23010131
Jeong YJ, Oh HK, Park SH, Bong JG. Prognostic Significance of Activated Leukocyte Cell Adhesion Molecule (ALCAM) in Association with Promoter Methylation of the ALCAM Gene in Breast Cancer. Molecules. 2018; 23(1):131. https://doi.org/10.3390/molecules23010131
Chicago/Turabian StyleJeong, Young Ju, Hoon Kyu Oh, Sung Hwan Park, and Jin Gu Bong. 2018. "Prognostic Significance of Activated Leukocyte Cell Adhesion Molecule (ALCAM) in Association with Promoter Methylation of the ALCAM Gene in Breast Cancer" Molecules 23, no. 1: 131. https://doi.org/10.3390/molecules23010131
APA StyleJeong, Y. J., Oh, H. K., Park, S. H., & Bong, J. G. (2018). Prognostic Significance of Activated Leukocyte Cell Adhesion Molecule (ALCAM) in Association with Promoter Methylation of the ALCAM Gene in Breast Cancer. Molecules, 23(1), 131. https://doi.org/10.3390/molecules23010131





