A Unique and Simple Approach to Improve Sensitivity in 15N-NMR Relaxation Measurements for NH3+ Groups: Application to a Protein-DNA Complex
Abstract
:1. Introduction
2. Results
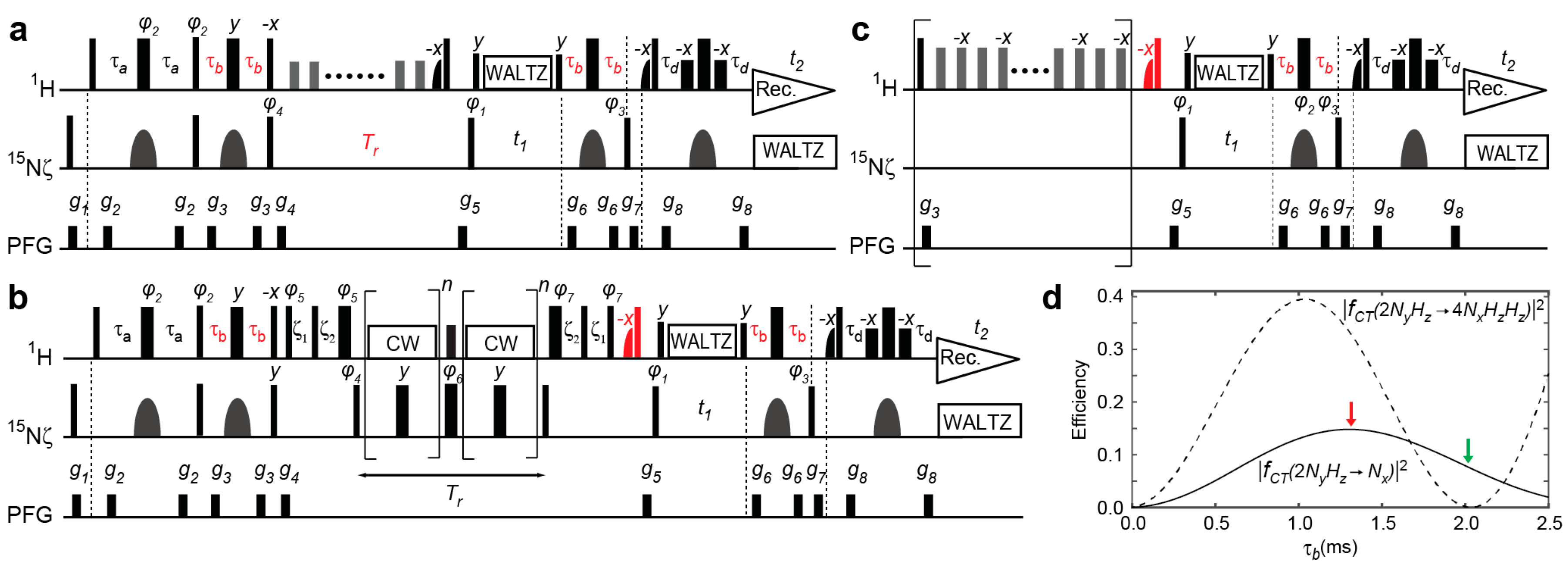
2.1. Previous and Current Approaches to Eliminating the Adverse Effects of Multi-Spin Order Terms
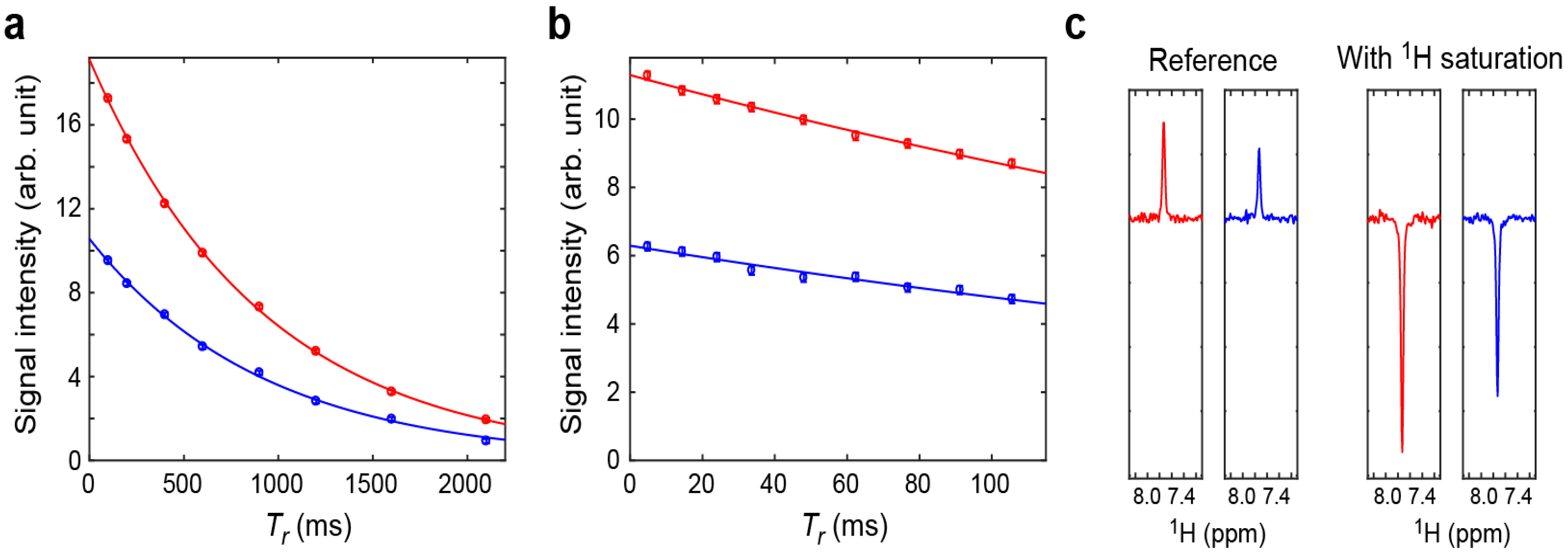
2.2. Assessment of the Sensitivity-Improved 15N R1 Experiment for NH3+ Groups
2.3. Assessment of the Sensitivity-Improved 15N R2 Experiment for NH3+ Groups
2.4. Assessment of the Sensitivity-Improved Heteronuclear NOE Experiment for NH3+ Groups
3. Discussion
4. Materials and Methods
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Boehr, D.D.; Dyson, H.J.; Wright, P.E. An NMR perspective on enzyme dynamics. Chem. Rev. 2006, 106, 3055–3079. [Google Scholar] [CrossRef] [PubMed]
- Clore, G.M.; Iwahara, J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009, 109, 4108–4139. [Google Scholar] [CrossRef] [PubMed]
- Kalodimos, C.G. NMR reveals novel mechanisms of protein activity regulation. Protein Sci. 2011, 20, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Kay, L.E. New views of functionally dynamic proteins by solution NMR spectroscopy. J. Mol. Biol. 2016, 428, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Loria, J.P.; Berlow, R.B.; Watt, E.D. Characterization of enzyme motions by solution NMR relaxation dispersion. Acc. Chem. Res. 2008, 41, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.G., III. NMR probes of molecular dynamics: Overview and comparison with other techniques. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 129–155. [Google Scholar] [CrossRef] [PubMed]
- Villali, J.; Kern, D. Choreographing an enzyme’s dance. Curr. Opin. Chem. Biol. 2010, 14, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Wand, A.J. The dark energy of proteins comes to light: Conformational entropy and its role in protein function revealed by NMR relaxation. Curr. Opin. Struct. Biol. 2013, 23, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Stafford, K.A.; Ferrage, F.; Cho, J.H.; Palmer, A.G., III. Side chain dynamics of carboxyl and carbonyl groups in the catalytic function of Escherichia coli ribonuclease H. J. Am. Chem. Soc. 2013, 135, 18024–18027. [Google Scholar] [CrossRef] [PubMed]
- Paquin, R.; Ferrage, F.; Mulder, F.A.; Akke, M.; Bodenhausen, G. Multiple-Timescale dynamics of side-chain carboxyl and carbonyl groups in proteins by 13C nuclear spin relaxation. J. Am. Chem. Soc. 2008, 130, 15805–15807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, A.L.; Kay, L.E. Quantifying millisecond time-scale exchange in proteins by CPMG relaxation dispersion NMR spectroscopy of side-chain carbonyl groups. J. Biomol. NMR 2011, 50, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Werbeck, N.D.; Kirkpatrick, J.; Hansen, D.F. Probing arginine side-chains and their dynamics with carbon-detected NMR spectroscopy: Application to the 42 kDa human histone deacetylase 8 at high pH. Angew. Chem. Int. Ed. Engl. 2013, 52, 3145–3147. [Google Scholar] [CrossRef] [PubMed]
- Trbovic, N.; Cho, J.H.; Abel, R.; Friesner, R.A.; Rance, M.; Palmer, A.G., III. Protein side-chain dynamics and residual conformational entropy. J. Am. Chem. Soc. 2009, 131, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.M.; Esadze, A.; Manoharan, M.; Brüschweiler, R.; Gorenstein, D.G.; Iwahara, J. Direct observation of the ion-pair dynamics at a protein-DNA interface by NMR spectroscopy. J. Am. Chem. Soc. 2013, 135, 3613–3619. [Google Scholar] [CrossRef] [PubMed]
- Esadze, A.; Li, D.W.; Wang, T.; Brüschweiler, R.; Iwahara, J. Dynamics of lysine side-chain amino groups in a protein studied by heteronuclear 1H-15N NMR spectroscopy. J. Am. Chem. Soc. 2011, 133, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Zandarashvili, L.; Esadze, A.; Iwahara, J. NMR studies on the dynamics of hydrogen bonds and ion pairs involving lysine side chains of proteins. Adv. Protein Chem. Struct. Biol. 2013, 93, 37–80. [Google Scholar] [PubMed]
- Zandarashvili, L.; Li, D.W.; Wang, T.; Brüschweiler, R.; Iwahara, J. Signature of mobile hydrogen bonding of lysine side chains from long-range 15N-13C scalar J-couplings and computation. J. Am. Chem. Soc. 2011, 133, 9192–9195. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Esadze, A.; Zandarashvili, L.; Nguyen, D.; Pettitt, B.M.; Iwahara, J. Dynamic equilibria of short-range electrostatic interactions at molecular interfaces of protein-DNA complexes. J. Phys. Chem. Lett. 2015, 6, 2733–2737. [Google Scholar] [CrossRef] [PubMed]
- Iwahara, J.; Esadze, A.; Zandarashvili, L. Physicochemical properties of ion pairs of biological macromolecules. Biomolecules 2015, 5, 2435–2463. [Google Scholar] [CrossRef] [PubMed]
- Esadze, A.; Chen, C.; Zandarashvili, L.; Roy, S.; Pettitt, B.M.; Iwahara, J. Changes in conformational dynamics of basic side chains upon protein-DNA association. Nucleic Acids Res. 2016, 44, 6961–6970. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Zandarashvili, L.; White, M.A.; Iwahara, J. Stereospecific effects of oxygen-to-sulfur substitution in DNA phosphate on ion pair dynamics and protein-DNA affinity. Chembiochem 2016, 17, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Zandarashvili, L.; Esadze, A.; Kemme, C.A.; Chattopadhyay, A.; Nguyen, D.; Iwahara, J. Residence times of molecular complexes in solution from NMR data of intermolecular hydrogen-bond scalar coupling. J. Phys. Chem. Lett. 2016, 7, 820–824. [Google Scholar] [CrossRef] [PubMed]
- Zandarashvili, L.; Esadze, A.; Vuzman, D.; Kemme, C.A.; Levy, Y.; Iwahara, J. Balancing between affinity and speed in target DNA search by zinc-finger proteins via modulation of dynamic conformational ensemble. Proc. Natl. Acad. Sci. USA 2015, 112, E5142–E5149. [Google Scholar] [CrossRef] [PubMed]
- Zandarashvili, L.; Iwahara, J. Temperature dependence of internal motions of protein side-chain NH3+ groups: Insight into energy barriers for transient breakage of hydrogen bonds. Biochemistry 2015, 54, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Zandarashvili, L.; Nguyen, D.; Anderson, K.M.; White, M.A.; Gorenstein, D.G.; Iwahara, J. Entropic enhancement of protein-DNA affinity by oxygen-to-sulfur substitution in DNA phosphate. Biophys. J. 2015, 109, 1026–1037. [Google Scholar] [CrossRef] [PubMed]
- Iwahara, J.; Jung, Y.S.; Clore, G.M. Heteronuclear NMR spectroscopy for lysine NH3 groups in proteins: Unique effect of water exchange on 15N transverse relaxation. J. Am. Chem. Soc. 2007, 129, 2971–2980. [Google Scholar] [CrossRef] [PubMed]
- Liepinsh, E.; Otting, G. Proton exchange rates from amino acid side chains—Implications for image contrast. Magn. Reson. Med. 1996, 35, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Segawa, T.; Kateb, F.; Duma, L.; Bodenhausen, G.; Pelupessy, P. Exchange rate constants of invisible protons in proteins determined by NMR spectroscopy. Chembiochem 2008, 9, 537–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abragam, A. Thermal relaxation in liquids and gases. In The Principle of Nuclear Magnetism; Carendon Press: Oxford, UK, 1961; pp. 264–353. [Google Scholar]
- Bax, A.; Ikura, M.; Kay, L.E.; Torchia, D.A.; Tschudin, R. Comparison of different modes of two-dimensional reverse-correlation NMR for the study of proteins. J. Magn. Reson. 1990, 86, 304–318. [Google Scholar] [CrossRef]
- Ollerenshaw, J.E.; Tugarinov, V.; Kay, L.E. Methyl TROSY: Explanation and experimental verification. Magn. Reson. Chem. 2003, 41, 843–852. [Google Scholar] [CrossRef]
- Esadze, A.; Zandarashvili, L.; Iwahara, J. Effective strategy to assign 1H-15N heteronuclear correlation NMR signals from lysine side-chain NH3+ groups of proteins at low temperature. J. Biomol. NMR 2014, 60, 23–27. [Google Scholar] [CrossRef]
- Poon, D.K.; Schubert, M.; Au, J.; Okon, M.; Withers, S.G.; McIntosh, L.P. Unambiguous determination of the ionization state of a glycoside hydrolase active site lysine by 1H-15N heteronuclear correlation spectroscopy. J. Am. Chem. Soc. 2006, 128, 15388–15389. [Google Scholar] [CrossRef] [PubMed]
- Wüthrich, K. NMR of Proteins and Nucleic Acids; Wiley-Interscience: New York, NY, USA, 1986; pp. 23–25. [Google Scholar]
- Sørensen, O.W.; Eich, G.W.; Levitt, M.H.; Bodenhausen, G.; Ernst, R.R. Product operator-formalism for the description of NMR pulse experiments. Prog. Nucl. Magn. Reson. Spectrosc. 1983, 16, 163–192. [Google Scholar] [CrossRef]
- Kupče, E.; Boyd, J.; Campbell, I.D. Short selective pulses for biochemical applications. J. Magn. Reson. Ser. B 1995, 106, 300–303. [Google Scholar] [CrossRef]
- Hansen, D.F.; Vallurupalli, P.; Kay, L.E. An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J. Phys. Chem. B 2008, 112, 5898–5904. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.F.; Kay, L.E. Improved magnetization alignment schemes for spin-lock relaxation experiments. J. Biomol. NMR 2007, 37, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Ernst, R.R.; Bodenhausen, G.; Wokaun, A. Heteronuclear polarization transfer. In Principles of Nuclear Magnetic Resonance in One and Two Dimensions; Oxford University Press: New York, NY, USA, 1987; pp. 180–201. [Google Scholar]
- Van de Ven, F.J.M. Dephasing coherences. In Multidimensional NMR in Liquids: Basic Principles and Experimental Methods; VCH Publishers: New York, NY, USA, 1995; pp. 211–224. [Google Scholar]
- Kay, L.E.; Bull, T.E.; Nicholson, L.K.; Griesinger, C.; Schwalbe, H.; Bax, A.; Torchia, D.A. The measurement of heteronuclear transverse relaxation-times in AX3 spin systems via polarization-transfer techniques. J. Magn. Reson. 1992, 100, 538–558. [Google Scholar] [CrossRef]
- Palmer, A.G.; Wright, P.E.; Rance, M. Measurement of relaxation-time constants for methyl-groups by proton-detected heteronuclear NMR spectroscopy. Chem. Phys. Lett. 1991, 185, 41–46. [Google Scholar] [CrossRef]
- Kumar, A.; Grace, R.C.R.; Madhu, P.K. Cross-Correlations in NMR. Prog. NMR Spect. 2000, 37, 191–319. [Google Scholar] [CrossRef]
- Anderson, K.M.; Nguyen, D.; Esadze, A.; Zandrashvili, L.; Gorenstein, D.G.; Iwahara, J. A chemical approach for site-specific identification of NMR signals from protein side-chain NH3+ groups forming intermolecular ion pairs in protein-nucleic acid complexes. J. Biomol. NMR 2015, 62, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Blevins, R.A. NMR view: A computer-program for the visualization and analysis of NMR data. J. Biomol. NMR 1994, 4, 603–614. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |

| Parameters | K46 | K55 | K57 | K58 |
|---|---|---|---|---|
| 15N R1 (s−1) b | 1.093 ± 0.013 | 0.637 ± 0.005 | 1.035 ± 0.004 | 0.363 ± 0.002 |
| 15N R1 (s−1) c | 1.081 ± 0.023 | 0.617 ± 0.008 | 1.037 ± 0.008 | 0.364 ± 0.003 |
| 15N R2,ini (s−1) b | 2.55 ± 0.10 | 1.76 ± 0.07 | 2.95 ± 0.04 | 1.20 ± 0.03 |
| 15N R2,ini (s−1) c | 2.74 ± 0.20 | 2.05 ± 0.12 | 2.76 ± 0.06 | 1.14 ± 0.06 |
| Heteronuclear NOE b | −2.44 ± 0.12 | −2.83 ± 0.10 | −2.54 ± 0.05 | −2.71 ± 0.05 |
| Heteronuclear NOE c | −2.53 ± 0.18 | −2.75 ± 0.13 | −2.60 ± 0.08 | −2.65 ± 0.07 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, D.; Lokesh, G.L.R.; Volk, D.E.; Iwahara, J. A Unique and Simple Approach to Improve Sensitivity in 15N-NMR Relaxation Measurements for NH3+ Groups: Application to a Protein-DNA Complex. Molecules 2017, 22, 1355. https://doi.org/10.3390/molecules22081355
Nguyen D, Lokesh GLR, Volk DE, Iwahara J. A Unique and Simple Approach to Improve Sensitivity in 15N-NMR Relaxation Measurements for NH3+ Groups: Application to a Protein-DNA Complex. Molecules. 2017; 22(8):1355. https://doi.org/10.3390/molecules22081355
Chicago/Turabian StyleNguyen, Dan, Ganesh L. R. Lokesh, David E. Volk, and Junji Iwahara. 2017. "A Unique and Simple Approach to Improve Sensitivity in 15N-NMR Relaxation Measurements for NH3+ Groups: Application to a Protein-DNA Complex" Molecules 22, no. 8: 1355. https://doi.org/10.3390/molecules22081355






