Application of Solution NMR to Structural Studies on α-Helical Integral Membrane Proteins
Abstract
:1. Introduction
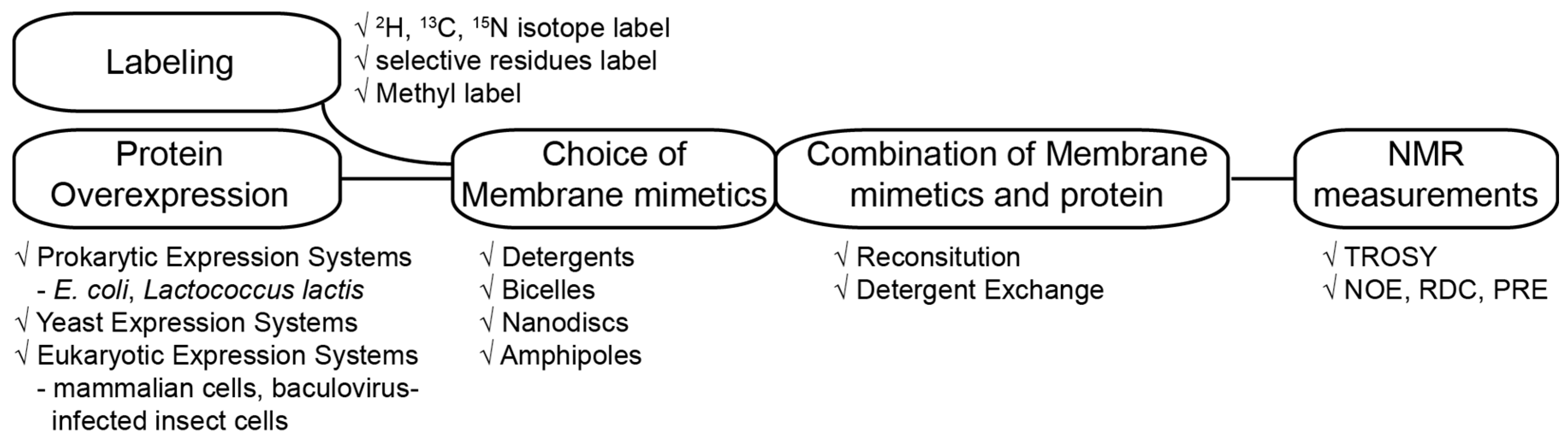
2. General Approaches to Structural Studies on Integral MPs
2.1. Protein Labeling
2.1.1. Protein Deuteration
2.1.2. Methyl Labeling
2.1.3. Amino Acid Type Selective Labeling
2.2. The Trends and Choice of Membrane-Mimetic Media for Structural Studies
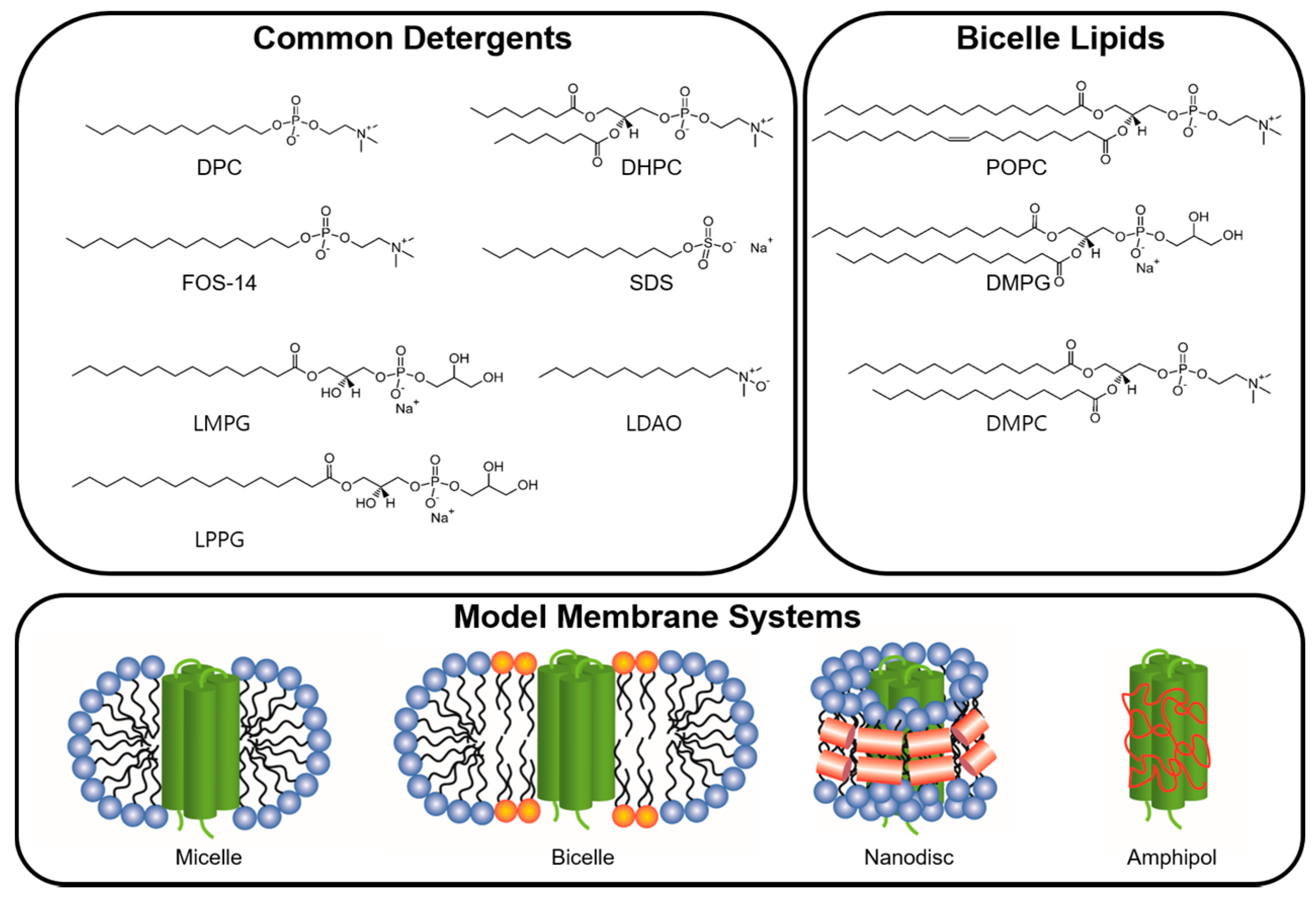
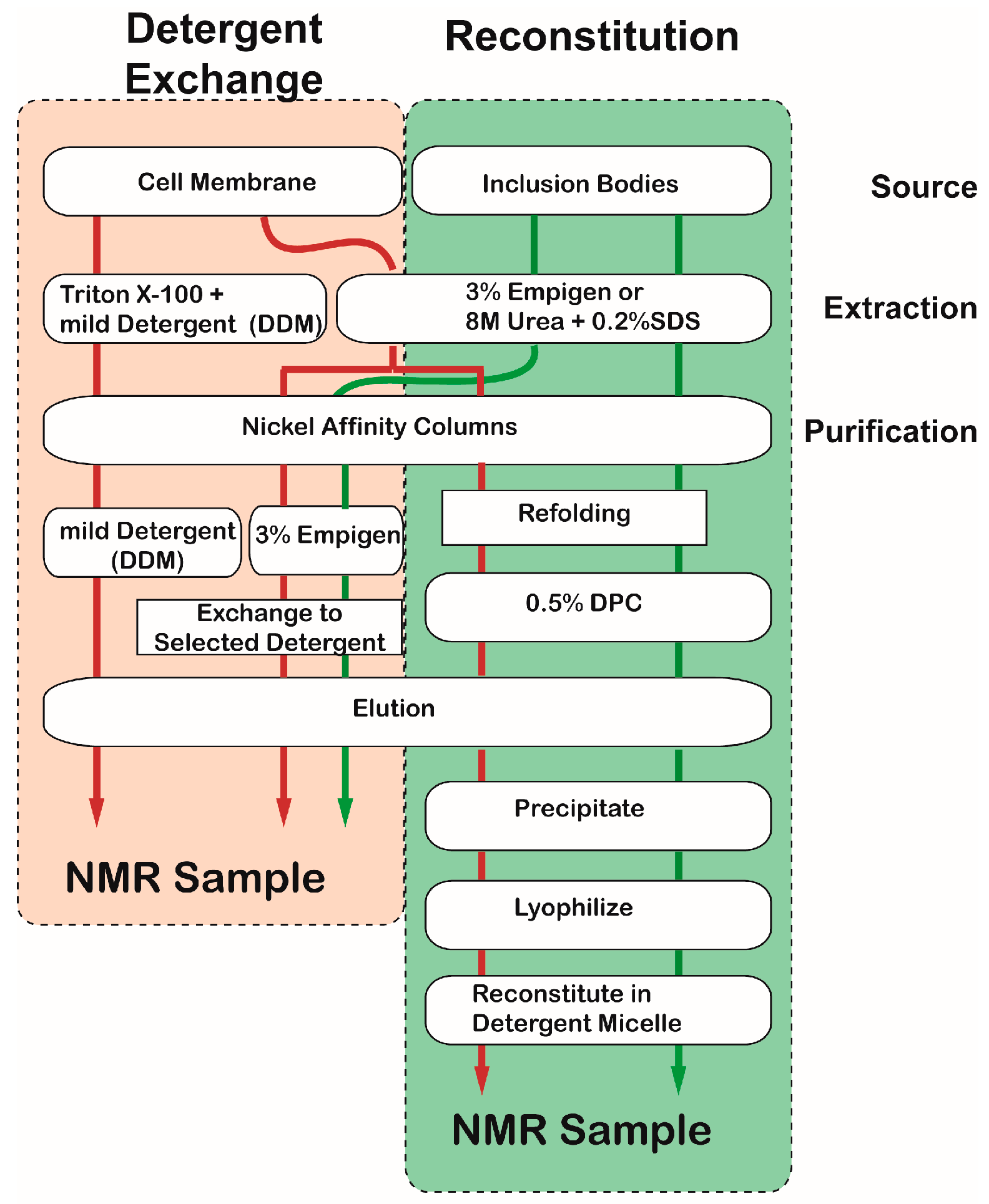
2.2.1. Detergents
2.2.2. Non-Micellar Media
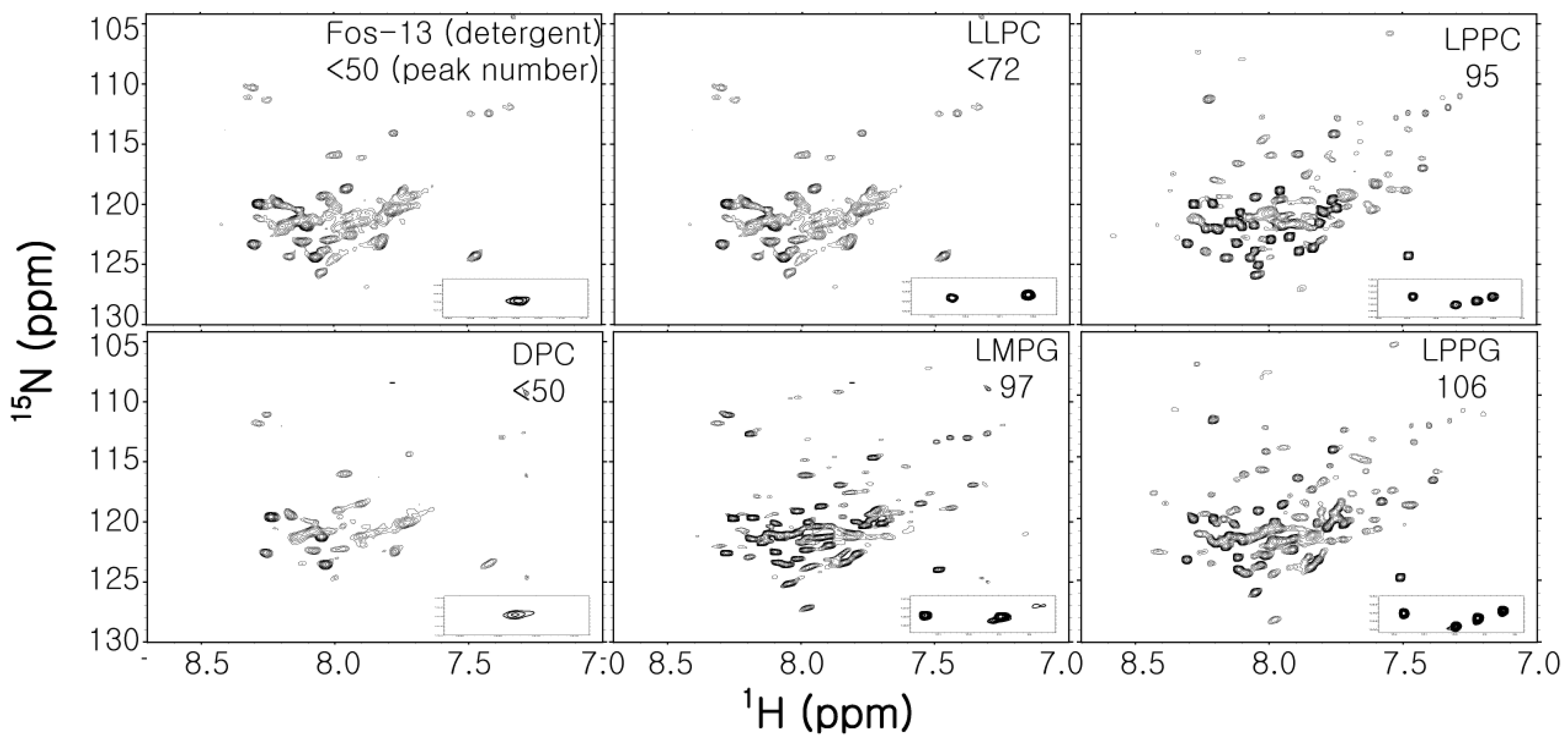
2.2.3. Analysis of Successful Membrane Mimetics
3. Illustration of Successful Structure Determination of Integral MPs
3.1. Structure of the Mitochondrial Translocator Protein in Complex with a Diagnostic Ligand
3.2. Unusual Architecture of the p7 Channel from Hepatitis C Virus
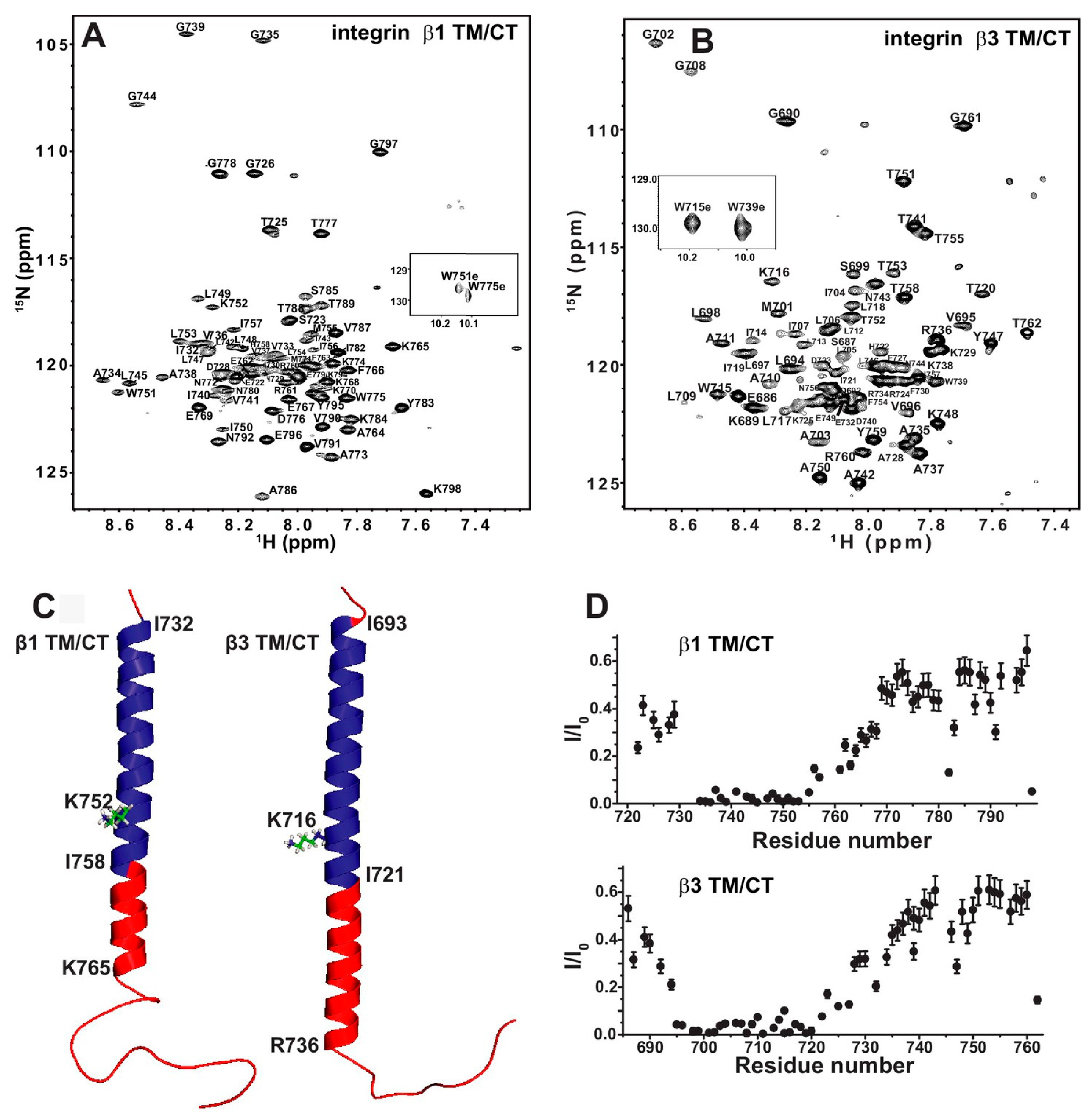
3.3. Implication of the Differing Roles of the β1 and β3 Tramsmembrane and Cytoplasmic Domains
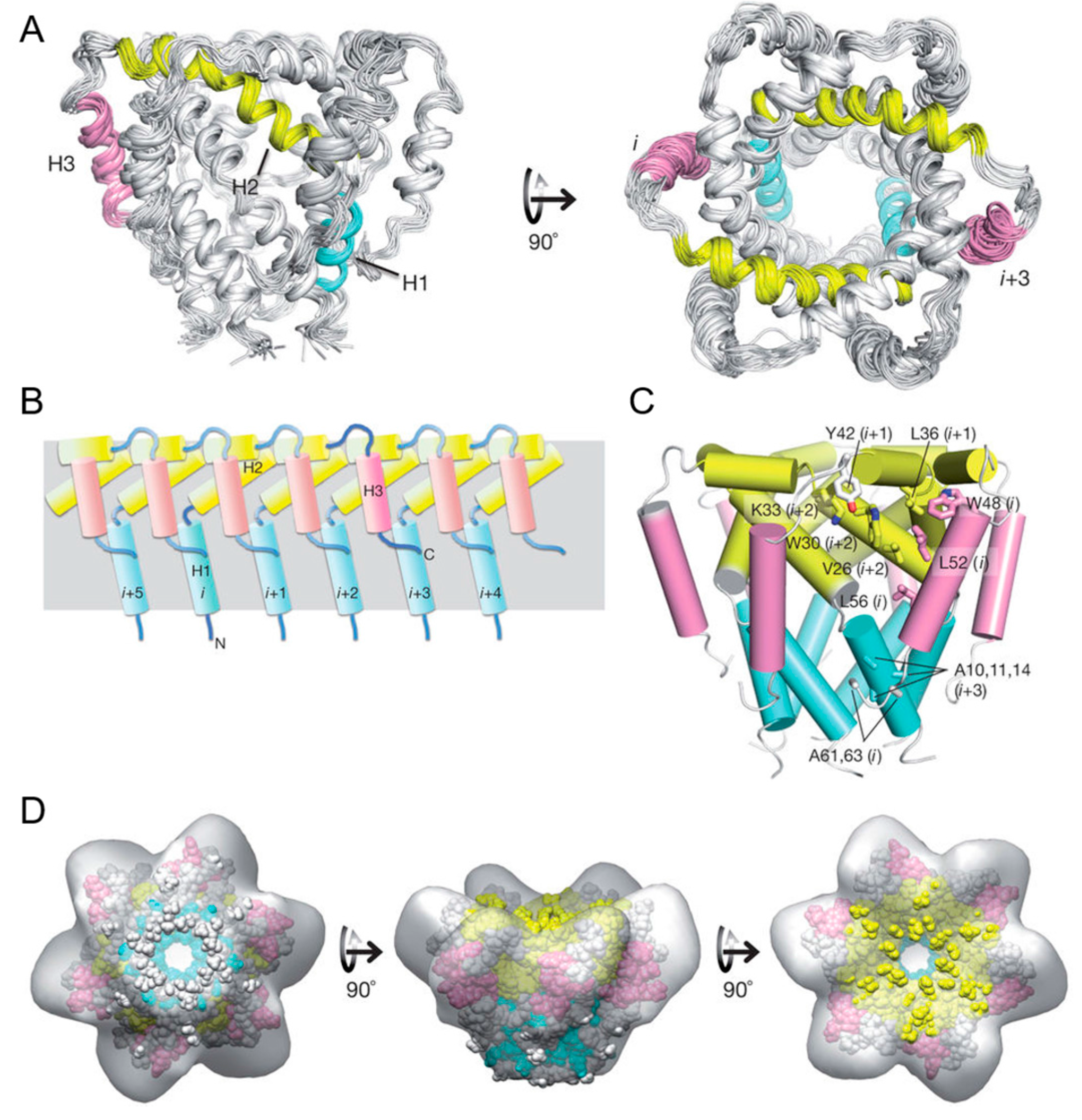
3.4. Architecture of the Mitochondrial Calcium Uniporter
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Almén, M.S.; Nordström, K.J.; Fredriksson, R.; Schiöth, H.B. Mapping the human membrane proteome: A majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Stroud, R.M. New tools in membrane protein determination. F1000 Biol. Rep. 2011, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Bertheleme, N.; Chae, P.S.; Singh, S.; Mossakowska, D.; Hann, M.M.; Smith, K.J.; Hubbard, J.A.; Dowell, S.J.; Byrne, B. Unlocking the secrets of the gatekeeper: Methods for stabilizing and crystallizing GPCRs. BBA Biomembr. 2013, 1828, 2583–2591. [Google Scholar] [CrossRef] [PubMed]
- Bukowska, M.A.; Grütter, M.G. New concepts and aids to facilitate crystallization. Curr. Opin. Struct. Biol. 2013, 23, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Weierstall, U.; James, D.; Wang, C.; White, T.A.; Wang, D.; Liu, W.; Spence, J.C.; Bruce Doak, R.; Nelson, G.; Fromme, P.; et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 2014, 5, 3309. [Google Scholar] [CrossRef] [PubMed]
- Van Horn, W.D.; Kim, H.J.; Ellis, C.D.; Hadziselimovic, A.; Sulistijo, E.S.; Karra, M.D.; Tian, C.; Sönnichsen, F.D.; Sanders, C.R. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science 2009, 324, 1726–1729. [Google Scholar] [CrossRef] [PubMed]
- Dev, J.; Park, D.; Fu, Q.; Chen, J.; Ha, H.J.; Ghantous, F.; Herrmann, T.; Chang, W.; Liu, Z.; Frey, G.; et al. Structural basis for membrane anchoring of HIV-1 envelope spike. Science 2016, 353, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Oxenoid, K.; Dong, Y.; Cao, C.; Cui, T.; Sancak, Y.; Markhard, A.L.; Grabarek, Z.; Kong, L.; Liu, Z.; Ouyang, B.; et al. Architecture of the mitochondrial calcium uniporter. Nature 2016, 533, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Reckel, S.; Gottstein, D.; Stehle, J.; Löhr, F.; Verhoefen, M.; Takeda, M.; Silvers, R.; Kainosho, M.; Glaubitz, C.; Wachtveitl, J.; et al. Solution NMR structure of proteorhodopsin. Angew. Chem. Int. Ed. 2011, 50, 11942–11946. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Bulkley, D.; Guo, Y.; Zhang, W.; Guo, Z.; Huynh, W.; Wu, S.; Meltzer, S.; Cheng, T.; Jan, L.Y.; et al. Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature 2017, 547, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Zhao, X.; Cao, P.; Lei, J.; Yan, N.; Gong, X. Structure of the Human Lipid Exporter ABCA1. Cell 2017, 169, 1228–1239.e10. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Das, B.B.; Casagrande, F.; Tian, Y.; Nothnagel, H.J.; Chu, M.; Kiefer, H.; Maier, K.; De Angelis, A.A.; Marassi, F.M.; et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature 2012, 491, 779–783. [Google Scholar] [PubMed]
- Opella, S.J.; Marassi, F.M. Applications of NMR to membrane proteins. Arch. Biochem. Biophys. 2017, 628, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Machius, M.; Dulubova, I.; Dai, H.; Sudhof, T.C.; Tomchick, D.R.; Rizo, J. Structural basis for a Munc13-1 homodimer to Munc13-1/RIM heterodimer switch. PLoS Biol. 2006, 4, e192. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Feng, W.; Wei, Z.; Miyanoiri, Y.; Wen, W.; Zhao, Y.; Zhang, M. Myosin VI undergoes cargo-mediated dimerization. Cell 2009, 138, 537–548. [Google Scholar] [CrossRef] [PubMed]
- OuYang, B.; Xie, S.; Berardi, M.J.; Zhao, X.; Dev, J.; Yu, W.; Sun, B.; Chou, J.J. Unusual architecture of the p7 channel from hepatitis C virus. Nature 2013, 498, 521–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Call, M.E.; Chou, J.J. A view into the blind spot: Solution NMR provides new insights into signal transduction across the lipid bilayer. Structure 2010, 18, 1559–1569. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Howell, S.C.; Van Horn, W.D.; Jeon, Y.H.; Sanders, C.R. Recent Advances in the Application of Solution NMR Spectroscopy to Multi-Span Integral Membrane Proteins. Prog. Nucl. Mag. Res. Spectrosc. 2009, 55, 335–360. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, N.; Malmendal, A.; Vosegaard, T. Techniques and applications of NMR to membrane proteins. Mol. Membr. Biol. 2004, 21, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Gautier, A. Structure determination of alpha-helical membrane proteins by solution-state NMR: Emphasis on retinal proteins. Biochim. Biophys. Acta 2014, 1837, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.; Pinto, C.; Houben, K.; Baldus, M. Nuclear magnetic resonance (NMR) applied to membrane-protein complexes. Q. Rev. Biophys. 2016, 49, e15. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Li, Q. Solution NMR study of integral membrane proteins. Curr. Opin. Chem. Biol. 2011, 15, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, S.; Hornak, V.; Yan, E.C.; Syrett, N.; Goncalves, J.A.; Hirshfeld, A.; Ziliox, M.; Sakmar, T.P.; Sheves, M.; Reeves, P.J.; et al. Helix movement is coupled to displacement of the second extracellular loop in rhodopsin activation. Nat. Struct. Mol. Biol. 2009, 16, 168–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goncalves, J.A.; South, K.; Ahuja, S.; Zaitseva, E.; Opefi, C.A.; Eilers, M.; Vogel, R.; Reeves, P.J.; Smith, S.O. Highly conserved tyrosine stabilizes the active state of rhodopsin. Proc. Natl. Acad. Sci. USA 2010, 107, 19861–19866. [Google Scholar] [CrossRef] [PubMed]
- Salom, D.; Cao, P.; Yuan, Y.; Miyagi, M.; Feng, Z.; Palczewski, K. Isotopic labeling of mammalian G protein-coupled receptors heterologously expressed in Caenorhabditis elegans. Anal. Biochem. 2015, 472, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.; Wider, G. TROSY in NMR studies of the structure and function of large biological macromolecules. Curr. Opin. Struct. Biol. 2003, 13, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Gardner, K.H.; Kay, L.E. The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 357–406. [Google Scholar] [CrossRef] [PubMed]
- Goto, N.K.; Kay, L.E. New developments in isotope labeling strategies for protein solution NMR spectroscopy. Curr. Opin. Struct. Biol. 2000, 10, 585–592. [Google Scholar] [CrossRef]
- Nietlispach, D.; Clowes, R.T.; Broadhurst, R.W.; Ito, Y.; Keeler, J.; Kelly, M.; Ashurst, J.; Oschkinat, H.; Domaille, P.J.; Laue, E.D. An approach to the structure determination of larger proteins using triple resonance NMR experiments in conjunction with random fractional deuteration. J. Am. Chem. Soc. 1996, 118, 407–415. [Google Scholar] [CrossRef]
- Sanders, C.R.; Sönnichsen, F. Solution NMR of membrane proteins: Practice and challenges. Magn. Reson. Chem. 2006, 44, S24–S40. [Google Scholar] [CrossRef] [PubMed]
- Gamble, T.R.; Clauser, K.R.; Kossiakoff, A.A. The production and X-ray structure determination of perdeuterated Staphylococcal nuclease. Biophys. Chem. 1994, 53, 15–25. [Google Scholar] [CrossRef]
- Liu, X.; Hanson, B.L.; Langan, P.; Viola, R.E. The effect of deuteration on protein structure: A high-resolution comparison of hydrogenous and perdeuterated haloalkane dehalogenase. Acta Crystallogr. D Biol. Crystallogr. 2007, 63, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cierpicki, T.; Jimenez, R.H.; Lukasik, S.M.; Ellena, J.F.; Cafiso, D.S.; Kadokura, H.; Beckwith, J.; Bushweller, J.H. NMR solution structure of the integral membrane enzyme DsbB: Functional insights into DsbB-catalyzed disulfide bond formation. Mol. Cell 2008, 31, 896–908. [Google Scholar] [CrossRef] [PubMed]
- Joh, N.H.; Min, A.; Faham, S.; Whitelegge, J.P.; Yang, D.; Woods, V.L.; Bowie, J.U. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature 2008, 453, 1266–1270. [Google Scholar] [CrossRef] [PubMed]
- Tugarinov, V.; Kay, L.E. Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. Chembiochem. Eur. J. Chem. Biol. 2005, 6, 1567–1577. [Google Scholar] [CrossRef] [PubMed]
- Janin, J.; Miller, S.; Chothia, C. Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol. 1988, 204, 155–164. [Google Scholar] [CrossRef]
- Nicholson, L.K.; Kay, L.E.; Baldisseri, D.M.; Arango, J.; Young, P.E.; Bax, A.; Torchia, D.A. Dynamics of methyl groups in proteins as studied by proton-detected 13C NMR spectroscopy. Application to the leucine residues of staphylococcal nuclease. Biochemistry (Mosc) 1992, 31, 5253–5263. [Google Scholar] [CrossRef]
- Tugarinov, V.; Hwang, P.M.; Kay, L.E. Nuclear magnetic resonance spectroscopy of high-molecular-weight proteins. Annu. Rev. Biochem. 2004, 73, 107–146. [Google Scholar] [CrossRef] [PubMed]
- Rosen, M.K.; Gardner, K.H.; Willis, R.C.; Parris, W.E.; Pawson, T.; Kay, L.E. Selective methyl group protonation of perdeuterated proteins. J. Mol. Biol. 1996, 263, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.O.; Ito, Y.; Raine, A.; Teichmann, S.; Ben-Tovim, L.; Nietlispach, D.; Broadhurst, R.W.; Terada, T.; Kelly, M.; Oschkinat, H.; et al. An approach to global fold determination using limited NMR data from larger proteins selectively protonated at specific residue types. J. Biomol. NMR 1996, 8, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Kerfah, R.; Plevin, M.J.; Sounier, R.; Gans, P.; Boisbouvier, J. Methyl-specific isotopic labeling: A molecular tool box for solution NMR studies of large proteins. Curr. Opin. Struct. Biol. 2015, 32, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Torizawa, T.; Shimizu, M.; Taoka, M.; Miyano, H.; Kainosho, M. Efficient production of isotopically labeled proteins by cell-free synthesis: A practical protocol. J. Biomol. NMR 2004, 30, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.T.; Sperling, L.J.; Frericks Schmidt, H.L.; Tang, M.; Samoilova, R.I.; Kumasaka, T.; Iwasaki, T.; Dikanov, S.A.; Rienstra, C.M.; Gennis, R.B. A rapid and robust method for selective isotope labeling of proteins. Methods 2011, 55, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.W.; Perez, J.A.; Ferguson, S.J.; Campbell, I.D. The specific incorporation of labelled aromatic amino acids into proteins through growth of bacteria in the presence of glyphosate. Application to fluorotryptophan labelling to the H(+)-ATPase of Escherichia coli and NMR studies. FEBS Lett. 1990, 272, 34–36. [Google Scholar] [CrossRef]
- O'Grady, C.; Rempel, B.L.; Sokaribo, A.; Nokhrin, S.; Dmitriev, O.Y. One-step amino acid selective isotope labeling of proteins in prototrophic Escherichia coli strains. Anal. Biochem. 2012, 426, 126–128. [Google Scholar] [CrossRef] [PubMed]
- Tong, K.I.; Yamamoto, M.; Tanaka, T. A simple method for amino acid selective isotope labeling of recombinant proteins in E. coli. J. Biomol. NMR 2008, 42, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Rosen, M.J.; Kunjappu, J.T. Surfactants and Interfacial Phenomena, 4th ed.; John Wiley & Sons: New York, NY, USA, 2012; pp. 123–191. ISBN 9780470541944. [Google Scholar]
- Tanford, C. The Hydrophobic Effect: Formation of Micelles and Biological Membranes, 2nd ed.; J. Wiley.: New York, NY, USA, 1980; pp. 60–123. ISBN 0471048933. [Google Scholar]
- Helenius, A.; McCaslin, D.R.; Fries, E.; Tanford, C. Properties of detergents. Method Enzymol. 1979, 56, 734–749. [Google Scholar]
- Neugebauer, J.M. Detergents: An overview. Methods Enzymol. 1990, 182, 239–253. [Google Scholar] [PubMed]
- Lipfert, J.; Columbus, L.; Chu, V.B.; Lesley, S.A.; Doniach, S. Size and shape of detergent micelles determined by small-angle X-ray scattering. J. Phys. Chem. B 2007, 111, 12427–12438. [Google Scholar] [CrossRef] [PubMed]
- le Maire, M.; Champeil, P.; Moller, J.V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 2000, 1508, 86–111. [Google Scholar] [CrossRef]
- Stafford, R.E.; Fanni, T.; Dennis, E.A. Interfacial properties and critical micelle concentration of lysophospholipids. Biochemistry (Mosc) 1989, 28, 5113–5120. [Google Scholar] [CrossRef]
- Chou, J.J.; Baber, J.L.; Bax, A. Characterization of phospholipid mixed micelles by translational diffusion. J. Biomol. NMR 2004, 29, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Lin, F. Social and economic factors affecting China fertility transition. Renkou Yanjiu 1987, 15–21. [Google Scholar]
- Strop, P.; Brunger, A.T. Refractive index-based determination of detergent concentration and its application to the study of membrane proteins. Protein Sci. 2005, 14, 2207–2211. [Google Scholar] [CrossRef] [PubMed]
- Green, M.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012; pp. 1481–1636. ISBN 1936113422. [Google Scholar]
- Kim, J.H.; Schlebach, J.P.; Lu, Z.; Peng, D.; Reasoner, K.C.; Sanders, C.R. A pH-Mediated Topological Switch within the N-Terminal Domain of Human Caveolin-3. Biophys. J. 2016, 110, 2475–2485. [Google Scholar] [CrossRef] [PubMed]
- Harbuz, M.S.; Richards, L.J.; Chover-Gonzalez, A.J.; Marti-Sistac, O.; Jessop, D.S. Stress in autoimmune disease models. Ann. N. Y. Acad. Sci. 2006, 1069, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Columbus, L.; Lipfert, J.; Jambunathan, K.; Fox, D.A.; Sim, A.Y.; Doniach, S.; Lesley, S.A. Mixing and matching detergents for membrane protein NMR structure determination. J. Am. Chem. Soc. 2009, 131, 7320–7326. [Google Scholar] [CrossRef] [PubMed]
- Tate, C.G. Practical considerations of membrane protein instability during purification and crystallisation. Methods Mol. Biol. 2010, 601, 187–203. [Google Scholar] [PubMed]
- Schmidt, T.; Ye, F.; Situ, A.J.; An, W.; Ginsberg, M.H.; Ulmer, T.S. A Conserved Ectodomain-Transmembrane Domain Linker Motif Tunes the Allosteric Regulation of Cell Surface Receptors. J. Biol. Chem. 2016, 291, 17536–17546. [Google Scholar] [CrossRef] [PubMed]
- Kroncke, B.M.; Van Horn, W.D.; Smith, J.; Kang, C.; Welch, R.C.; Song, Y.; Nannemann, D.P.; Taylor, K.C.; Sisco, N.J.; George, A.L., Jr.; et al. Structural basis for KCNE3 modulation of potassium recycling in epithelia. Sci. Adv. 2016, 2, e1501228. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Fu, T.M.; Cruz, A.C.; Sengupta, P.; Thomas, S.K.; Wang, S.; Siegel, R.M.; Wu, H.; Chou, J.J. Structural Basis and Functional Role of Intramembrane Trimerization of the Fas/CD95 Death Receptor. Mol. Cell 2016, 61, 602–613. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Mittendorf, K.F.; Lu, Z.; Sanders, C.R. Impact of bilayer lipid composition on the structure and topology of the transmembrane amyloid precursor C99 protein. J. Am. Chem. Soc. 2014, 136, 4093–4096. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, T.K.; Grinkova, Y.V.; Bayburt, T.H.; Denisov, I.G.; Zolnerciks, J.K.; Atkins, W.M.; Sligar, S.G. Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Method. Enzymol. 2009, 464, 211–231. [Google Scholar]
- Puthenveetil, R.; Nguyen, K.; Vinogradova, O. Nanodiscs and Solution NMR: Preparation, application and challenges. Nanotechnol. Rev. 2017, 6, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Tzitzilonis, C.; Eichmann, C.; Maslennikov, I.; Choe, S.; Riek, R. Detergent/nanodisc screening for high-resolution NMR studies of an integral membrane protein containing a cytoplasmic domain. PLoS ONE 2013, 8, e54378. [Google Scholar] [CrossRef] [PubMed]
- Raschle, T.; Hiller, S.; Etzkorn, M.; Wagner, G. Nonmicellar systems for solution NMR spectroscopy of membrane proteins. Curr. Opin. Struct. Biol. 2010, 20, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Bibow, S.; Carneiro, M.G.; Sabo, T.M.; Schwiegk, C.; Becker, S.; Riek, R.; Lee, D. Measuring membrane protein bond orientations in nanodiscs via residual dipolar couplings. Protein Sci. 2014, 23, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Denisov, I.G.; Grinkova, Y.V.; Lazarides, A.A.; Sligar, S.G. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 2004, 126, 3477–3487. [Google Scholar] [CrossRef] [PubMed]
- Popot, J.L. Amphipols, nanodiscs, and fluorinated surfactants: Three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu. Rev. Biochem. 2010, 79, 737–775. [Google Scholar] [CrossRef] [PubMed]
- Champeil, P.; Menguy, T.; Tribet, C.; Popot, J.L.; le Maire, M. Interaction of amphipols with sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 2000, 275, 18623–18637. [Google Scholar] [CrossRef] [PubMed]
- Picard, M.; Duval-Terrie, C.; De, E.; Champeil, P. Stabilization of membranes upon interaction of amphipathic polymers with membrane proteins. Protein Sci. 2004, 13, 3056–3058. [Google Scholar] [CrossRef] [PubMed]
- Tribet, C.; Audebert, R.; Popot, J.L. Amphipols: Polymers that keep membrane proteins soluble in aqueous solutions. Proc. Natl. Acad. Sci. USA. 1996, 93, 15047–15050. [Google Scholar] [CrossRef] [PubMed]
- Planchard, N.; Point, E.; Dahmane, T.; Giusti, F.; Renault, M.; Le Bon, C.; Durand, G.; Milon, A.; Guittet, E.; Zoonens, M.; et al. The use of amphipols for solution NMR studies of membrane proteins: Advantages and constraints as compared to other solubilizing media. J. Membr. biol. 2014, 247, 827–842. [Google Scholar] [CrossRef] [PubMed]
- Catoire, L.J.; Damian, M.; Giusti, F.; Martin, A.; van Heijenoort, C.; Popot, J.L.; Guittet, E.; Baneres, J.L. Structure of a GPCR ligand in its receptor-bound state: Leukotriene B4 adopts a highly constrained conformation when associated to human BLT2. J. Am. Chem. Soc. 2010, 132, 9049–9057. [Google Scholar] [CrossRef] [PubMed]
- Nasr, M.L.; Baptista, D.; Strauss, M.; Sun, Z.J.; Grigoriu, S.; Huser, S.; Pluckthun, A.; Hagn, F.; Walz, T.; Hogle, J.M.; et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods 2017, 14, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Jaremko, L.; Jaremko, M.; Giller, K.; Becker, S.; Zweckstetter, M. Structure of the mitochondrial translocator protein in complex with a diagnostic ligand. Science 2014, 343, 1363–1366. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, V.; Baraldi, M.; Guilarte, T.R.; Knudsen, T.B.; Lacapere, J.J.; Lindemann, P.; Norenberg, M.D.; Nutt, D.; Weizman, A.; Zhang, M.R.; et al. Translocator protein (18kDa): New nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci. 2006, 27, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Rupprecht, R.; Papadopoulos, V.; Rammes, G.; Baghai, T.C.; Fan, J.; Akula, N.; Groyer, G.; Adams, D.; Schumacher, M. Translocator protein (18 kDa) (TSPO) as a therapeutic target for neurological and psychiatric disorders. Nat. Rev. Drug Discov. 2010, 9, 971–988. [Google Scholar] [CrossRef] [PubMed]
- Braestrup, C.; Squires, R.F. Specific benzodiazepine receptors in rat brain characterized by high-affinity (3H)diazepam binding. Proc. Natl. Acad. Sci. USA 1977, 74, 3805–3809. [Google Scholar] [CrossRef] [PubMed]
- Veenman, L.; Papadopoulos, V.; Gavish, M. Channel-like functions of the 18-kDa translocator protein (TSPO): Regulation of apoptosis and steroidogenesis as part of the host-defense response. Curr. Pharm. Des. 2007, 13, 2385–2405. [Google Scholar] [CrossRef] [PubMed]
- Veenman, L.; Gavish, M. The role of 18 kDa mitochondrial translocator protein (TSPO) in programmed cell death, and effects of steroids on TSPO expression. Curr. Mol. Med. 2012, 12, 398–412. [Google Scholar] [CrossRef] [PubMed]
- Frank, W.; Baar, K.M.; Qudeimat, E.; Woriedh, M.; Alawady, A.; Ratnadewi, D.; Gremillon, L.; Grimm, B.; Reski, R. A mitochondrial protein homologous to the mammalian peripheral-type benzodiazepine receptor is essential for stress adaptation in plants. Plant J. 2007, 51, 1004–1018. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.R.; Matthews, P.M. Imaging brain microglial activation using positron emission tomography and translocator protein-specific radioligands. Int. Rev. Neurobiol. 2011, 101, 19–39. [Google Scholar] [PubMed]
- Guntert, P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar] [PubMed]
- Schwieters, C.D.; Kuszewski, J.J.; Tjandra, N.; Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003, 160, 65–73. [Google Scholar] [CrossRef]
- Deisenhofer, J.; Michel, H. The Photosynthetic Reaction Center from the Purple Bacterium Rhodopseudomonas viridis. Science 1989, 245, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Almeida, F.C.; Opella, S.J. fd coat protein structure in membrane environments: Structural dynamics of the loop between the hydrophobic trans-membrane helix and the amphipathic in-plane helix. J. Mol. Biol. 1997, 270, 481–495. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Papadopoulos, V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology 1998, 139, 4991–4997. [Google Scholar] [CrossRef] [PubMed]
- Jamin, N.; Neumann, J.M.; Ostuni, M.A.; Vu, T.K.; Yao, Z.X.; Murail, S.; Robert, J.C.; Giatzakis, C.; Papadopoulos, V.; Lacapere, J.J. Characterization of the cholesterol recognition amino acid consensus sequence of the peripheral-type benzodiazepine receptor. Mol. Endocrinol. 2005, 19, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Mathew, S.; Chen, J.; Hadziselimovic, A.; Palamuttam, R.; Hudson, B.G.; Fassler, R.; Pozzi, A.; Sanders, C.R.; Zent, R. Implications of the differing roles of the beta1 and beta3 transmembrane and cytoplasmic domains for integrin function. eLife 2016, 5, e18633. [Google Scholar] [CrossRef] [PubMed]
- Kamer, K.J.; Mootha, V.K. The molecular era of the mitochondrial calcium uniporter. Nat. Rev. Mol. Cell Biol. 2015, 16, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Schnell, J.R.; Chou, J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 2008, 451, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Sancak, Y.; Markhard, A.L.; Kitami, T.; Kovacs-Bogdan, E.; Kamer, K.J.; Udeshi, N.D.; Carr, S.A.; Chaudhuri, D.; Clapham, D.E.; Li, A.A.; et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 2013, 342, 1379–1382. [Google Scholar] [CrossRef] [PubMed]
- Kovacs-Bogdan, E.; Sancak, Y.; Kamer, K.J.; Plovanich, M.; Jambhekar, A.; Huber, R.J.; Myre, M.A.; Blower, M.D.; Mootha, V.K. Reconstitution of the mitochondrial calcium uniporter in yeast. Proc. Natl. Acad. Sci. USA 2014, 111, 8985–8990. [Google Scholar] [CrossRef] [PubMed]



| Detergent | Charge | Molecular Weight | Critical Micelle Concentration (mM) | Aggregation Number | Reference |
|---|---|---|---|---|---|
| Dodecylphosphocholine (DPC) | Zwitterionic | 352 | 1.5 | 50–60 | [54] |
| Lyso-myristoyl phosphatidylglycerol (LMPG) | Anionic | 478 | 0.2–3 | ND | [55] |
| Dihexanoylphosphatidylcholine (DHPC) | Zwitterionic | 454 | 15 | 19–35 | [56,57] |
| Sodium dodecylsulfate (SDS) | Anionic | 288 | 1–7 | 62–101 | [54] |
| Lyso-palmitoyl phosphatidylglycerol (LPPG) | Anionic | 506 | 0.02–0.6 | 125 | [56] |
| N-Lauroyldimethyl amineoxide (LDAO) | Zwitterionic | 229 | 2 | 69–73 | [54] |
| β-Dodecylmaltoside (DDM) | Nonionic | 511 | 0.2 | 110-140 | [54] |
| n-Tetradecylphosphocholine (Fos-14) | Zwitterionic | 379.5 | 1.2 | 108 | [58] |
| Zwittergent 3–12 | Zwitterionic | 335.6 | 3 | 55–87 | [59] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sim, D.-W.; Lu, Z.; Won, H.-S.; Lee, S.-N.; Seo, M.-D.; Lee, B.-J.; Kim, J.-H. Application of Solution NMR to Structural Studies on α-Helical Integral Membrane Proteins. Molecules 2017, 22, 1347. https://doi.org/10.3390/molecules22081347
Sim D-W, Lu Z, Won H-S, Lee S-N, Seo M-D, Lee B-J, Kim J-H. Application of Solution NMR to Structural Studies on α-Helical Integral Membrane Proteins. Molecules. 2017; 22(8):1347. https://doi.org/10.3390/molecules22081347
Chicago/Turabian StyleSim, Dae-Won, Zhenwei Lu, Hyung-Sik Won, Seu-Na Lee, Min-Duk Seo, Bong-Jin Lee, and Ji-Hun Kim. 2017. "Application of Solution NMR to Structural Studies on α-Helical Integral Membrane Proteins" Molecules 22, no. 8: 1347. https://doi.org/10.3390/molecules22081347




