Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua
Abstract
1. Introduction
2. Results and Discussion
2.1. Characteristics of A. annua cpDNA
2.2. Long Repeat and SSR Analysis
2.3. Comparative Chloroplast Genomic Analysis
2.4. IR Contraction and Expansion in the A. annua cp Genome
2.5. Phylogenetic Analysis
3. Materials and Methods
3.1. DNA Sequencing, cp Genome Assembly, and Validation
3.2. Gene Annotation and Sequence Analyses
3.3. Genome Comparison
3.4. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Klayman, D.L. Qinghaosu (artemisinin): An antimalarial drug from China. Science 1985, 228, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Arrow, K.J.; Panosian, C.B.; Gelband, H. Saving Lives, Buying Time: Economics of Malaria Drugs in an Age of Resistance; National Academies Press: Washington, DC, USA, 2004. [Google Scholar]
- Tu, Y.Y. Artemisinin—A fift from traditional chinese medicine to the world (nobel lecture). Angew. Chem. Int. Ed. Engl. 2016, 55, 10210–10226. [Google Scholar] [CrossRef] [PubMed]
- Mert, A.; Krc, S.; Ayanoğlu, F. The effects of different plant densities on yield, yield components and quality of Artemisia annua L. Ecotypes. J. Herbs Spices Med. Plants 2002, 9, 413–418. [Google Scholar] [CrossRef]
- Delabays, N.; Simonnet, X.; Gaudin, M. The genetics of artemisinin content in Artemisia annua L. and the breeding of high yielding cultivars. Curr. Med. Chem. 2001, 8, 1795–1801. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.Y.; Zhou, H.R.; Lun, Y.; Hu, M.; Zhao, P.P. Studies on quality germplasm resources of Artemisia annua. Chin. Herbal Med. 1998, 29, 264–267. [Google Scholar]
- Hu, S.L.; Xu, Q.C.; Liu, J.F.; Gu, Y.X. Studies on plant resources of artemisinin. China J. Chin. Mater. Med. 1981, 2, 13–16. [Google Scholar]
- Guo, X.X.; Yang, X.Q.; Yang, R.Y. Salicylic acid and methyl jasmonate but not Rose Bengal enhance artemisinin production through invoking burst of endogenous singlet oxygen. Plant Sci. 2010, 178, 390–397. [Google Scholar] [CrossRef]
- Zeng, Q.P.; Zeng, X.M.; Yang, R.Y. Singlet oxygen as a signaling transducer for modulating artemisinin biosynthetic genes in Artemisia annua. Biol. Plantarum. 2011, 55, 69–674. [Google Scholar] [CrossRef]
- Sun, C.; Fan, C.; Zhang, F.; Niu, T.; Sun, Y.; Guo, X. Cloning and sequence analysis of ps1A1 and ps1A2 genes amplified specifically from the chloroplast and of maintainer of CMS Sorghum. Chin. J. Appl. Environ. Biol. 2003, 9, 501–505. [Google Scholar]
- Nielsen, A.Z.; Ziersen, B.; Jensen, K.; Lassne, L.M.; Olsen, C.E.; Moller, B.L.; Jensen, P.E. Redirecting photosynthetic reducing power toward bioactive natural product synthesis. ACS Synth. Biol. 2013, 2, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; Depamphilis, C.W.; Kai, F.M.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Mordent, C.W.; Ems, S.C.; Palmer, J.D. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: Loss or accelerated sequence evolution of tRNA and ribosomal protein genes. Mol. Evol. 1992, 35, 304–317. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.Q.; Raubeson, L.A.; Daniell, H.; dePamphilis, C.W.; Leebeans-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Qian, J.; Sun, Z.Y.; Xu, X.L.; Chen, S.L. Complete chloroplast genome of medicinal plant Lonicera japonica: Genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules 2017, 22, 249. [Google Scholar] [CrossRef] [PubMed]
- Soetaert, S.; Van Nieuwerburgh, F.; Brodelius, P.; Goossens, A.; Deforce, D. Transcriptome analysis of apical and sub-apical cells of Artemisia annua trichomes with next-generation-sequencing. In Proceedings of the 10th International Meeting on All Aspects of the Chemistry and Biology of Terpenes and Isoprenoids (Terpnet 2011): Biosynthesis and Function of Isoprenoids in Plants, Microorganisms and Parasites, Kalmar, Sweden, 22–26 May 2011; p. 170. [Google Scholar]
- Soetaert, S.S.; Neste, C.M.V.; Vandewoestyne, M.L.; Head, S.R.; Goossens, A.; Van Nieuwerburgh, F.C.; Deforce, D.L. Differential transcriptome analysis of glandular and filamentous trichomes in Artemisia annua. BMC Plant Biol. 2013, 13, 220. [Google Scholar] [CrossRef] [PubMed]
- Graham, I.A.; Besser, K.; Blumer, S.; Branigan, C.A.; Czechowski, T.; Elias, L.; Guterman, I.; Harvey, D.; Issac, P.G.; Khan, A.M.; et al. The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science 2010, 327, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Song, J.Y. Herbgenomics. China J. Chin. Mater. Med. 2016, 41, 3881–3889. [Google Scholar]
- Chen, S.L.; Song, J.Y.; Sun, C.; Xu, J.; Zhu, Y.J.; Verpoorte, R.; Fan, T.P. Herbal genomics: Examining the biology of traditional medicines. Science 2015, 347, 27–29. [Google Scholar]
- Nie, X.; Lv, S.; Zhang, Y.; Du, X.; Wang, L.; Biradar, S.S.; Tan, X.; Wan, F.; Weining, S. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PLoS ONE 2012, 7, e36869. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Shao, Y.; Li, Q.; Gao, J.; Zhang, R.; Lai, X.; Wang, D.; Zhang, H. The complete chloroplast genome sequence of the medicinal plant Andrographis paniculata. Mitochondr. DNA 2016, 27, 2347–2348. [Google Scholar]
- Jia, Y.; Yang, J.; He, Y.L.; He, Y.; Niu, C.; Gong, L.-L.; Li, Z.-H. Characterization of the whole chloroplast genome sequence of Acer davidii Franch (Aceraceae). Conserv. Genet. Resour. 2016, 8, 141–143. [Google Scholar] [CrossRef]
- Xiang, B.; Li, X.; Qian, J.; Wang, L.; Ma, L.; Tian, X.; Wang, Y. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii. Using the PacBio RS II platform. Molecules 2016, 21, 1029. [Google Scholar] [CrossRef] [PubMed]
- Clegg, M.T.; Gaut, B.S.; Learn, G.H.; Morton, B.R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 6795–6801. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huo, N.; Dong, L.; Wang, Y.; Zhang, S.; Yooung, H.A.; Feng, X.; Gu, Y.Q. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS ONE 2013, 8, e57533. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, E.; Takahashi, Y.; Lemieux, C.; Turmel, M.; Rochaix, J.D. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 1997, 16, 6095–6104. [Google Scholar] [CrossRef] [PubMed]
- Naver, H.; Boudreau, E.; Rochaix, J.D. Functional studies of Ycf3: Its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 2001, 13, 2731–2745. [Google Scholar] [CrossRef] [PubMed]
- Bryan, G.J.; McNicol, J.W.; Meyer, R.C.; Ramsay, G.; De Jong, W.S. Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants. Theor. Appl. Genet. 1999, 99, 859–867. [Google Scholar] [CrossRef]
- Provan, J. Novel chloroplast microsatellites reveal cytoplasmic variation in Arabidopsis thaliana. Mol. Ecol. 2000, 9, 2183–2185. [Google Scholar] [CrossRef] [PubMed]
- Flannery, M.L.; Mitchell, F.J.; Coyne, S.; Kavanagh, T.A.; Burke, J.I.; Salamin, N.; Dowding, P.; Hodkinson, T.R. Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs. Theor. Appl. Genet. 2006, 113, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Ebert, D.; Peakall, R. Chloroplast simple sequence repeats (cpSSRs): Technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Mol. Ecol. Resour. 2009, 9, 673–690. [Google Scholar] [CrossRef] [PubMed]
- Zhihai, H.; Jiang, X.; Shuiming, X.; Baosheng, L.; Yuan, G.; Chaochao, Z.; Xiaohui, Q.; Wen, X.; Shilin, C. Comparative optical genome analysis of two pangolin species: Manis pentadactyla and Manis javanica. Gigascience 2016, 5, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.H.; Zhao, Z.L.; Xu, H.X.; Chen, S.L.; Dorje, G. The complete chloroplast genome of Gentiana straminea (Gentianaceae), an endemic species to the Sino-Himalayan subregion. Gene 2016, 577, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boorem, J.L.; Jansen, R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007, 8, 174–201. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.J.; Cheng, C.L.; Chang, C.C.; Wu, C.L.; Su, T.M.; Chaw, S.M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008, 8, 36–50. [Google Scholar] [CrossRef] [PubMed]
- De Cambiaire, J.C.; Otis, C.; Lemieux, C.; Turmel, M. The complete chloroplast genome sequence of the chlorophycean green alga Scenedesmus obliquus reveals a compact gene organization and a biased distribution of genes on the two DNA strands. BMC Evol. Biol. 2006, 6, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kang, Y.; Shin, S.C.; Park, H.; Lee, H. Combined analysis of the chloroplast genome and transcriptome of the Antarctic vascular plant Deschampsia antarctica Desv. PLoS ONE 2014, 9, e92501. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yang, B.; Zhu, W.; Sun, L.; Tian, J.; Wang, X. The complete chloroplast genome sequence of Mahonia bealei (Berberidaceae) reveals a significant expansion of the inverted repeat and phylogenetic relationship with other angiosperms. Gene 2013, 528, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.J.; Choi, K.S.; Jansen, R.K. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol. 2005, 22, 1783–1792. [Google Scholar] [CrossRef] [PubMed]
- Timme, R.E.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am. J. Bot. 2007, 94, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Penaflor, C.; Kuehl, J.V.; Leebens-Mack, J.; Carlson, J.E.; dePamphilis, C.W.; Boore, J.L.; Jansen, R.K. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: Implications for the phylogenetic relationships of magnoliids. BMC Evol. Biol. 2006, 6, 77–97. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.R.; Dastidar, S.G.; Cai, Z.; Penaflor, C.; Kuehl, J.V.; Boore, J.L.; Janse, K. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol. Phylogenet. Evol. 2007, 45, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Raubeson, L.A.; Jansen, R.K. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science 1992, 255, 1697–1699. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Hahn, F.M.; Mcmahan, C.M.; Cornish, K.; Whalen, M.C. Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol. 2009, 9, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Palmer, J.D. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae). Proc. Natl. Acad. Sci. USA 1987, 84, 5818–5822. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Davis, J.I.; Soreng, R.J.; Garvin, D.; Anderson, M.J. Chloroplast DNA inversions and the origin of the grass family (Poaceae). Proc. Natl. Acad. Sci. USA 1992, 89, 7722–7726. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Nugent, J.M.; Herbon, L.A. Unusual structure of geranium chloroplast DNA: A triple-sized inverted repeat, extensive gene duplications, multiple inversions, and two repeat families. Proc. Natl. Acad. Sci. USA 1987, 84, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, Y.; Terachi, T.; Sasakuma, T. Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc. Natl. Acad. Sci. USA 1988, 85, 8573–8577. [Google Scholar] [CrossRef] [PubMed]
- Panero, J.L.; Funk, V.A. The value of sampling anomalous taxa in phylogenetic studies: Major clades of the Asteraceae revealed. Mol. Phylogenet. Evol. 2008, 47, 757–782. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, I.A.; Aguilar, J.F.; Panero, J.L.; Feliner, G.N. A phylogenetic analysis of Doronicum (Asteraceae, Senecioneae) based on morphological, nuclear ribosomal (ITS), and chloroplast (trnL-F) evidence. Mol. Phylogenet. Evol. 2001, 20, 41–64. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.B.; Peng, Y.; Chen, S.L.; Xiao, P.G. Introduction of Pharmaphylogeny. Mod. Tradit. Chin. Med. Mater. Med. World Sci. Technol. 2005, 7, 97–103. [Google Scholar]
- Shi, Q.H.; Yao, Z.P.; Zhang, H.; Xu, L.; Dai, P.H. Comparison of four methods of DNA extraction from Chickpea. J. Xinjiang Agric. Univ. 2009, 1, 64–67. [Google Scholar]
- Urreizti, R.; Garcia-Giralt, N.; Riancho, J.A.; Gibzakez-Macias, J.; Civit, S.; Guerris, R.; Yoskovitz, G.; Sarrion, P.; Mellivobsky, L.; Diez-Perez, A.; et al. COL1A1, haplotypes and hip fracture. J. Bone Miner. Res. 2012, 27, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Wang, L.; Cui, L.; Feng, K.; Liu, F.; Du, X.; Tong, W.; Niu, X.; Ji, W.; Weining, S. Global identification of MicroRNAs and their targets in barley under salinity stress. PLoS ONE 2015, 10, e0137990. [Google Scholar] [CrossRef] [PubMed]
- Gogniashvili, M.; Naskidashvili, P.; Bedoshvili, D.; Kotorashcili, N.; Kotaria, N.; Beridze, T. Complete chloroplast DNA sequences of Zanduri wheat (Triticum, spp.). Genet. Resour. Crop Evol. 2015, 62, 1269–1277. [Google Scholar] [CrossRef]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Acemel, R.D.; Tena, J.J.; Irastorzaazcarate, I.; Marletaz, F.; Comez-Marin, C.; de la Calle-Mustienes, E.; Bertrand, S.; Diaz, S.G.; Aldea, D.; Aury, J.M.; et al. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat. Genet. 2016, 48, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 715. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. Organellar Genome DRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Dubchak, I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.M.; Sun, J.T.; Xue, X.F.; Zhu, W.C.; Hong, X.Y. Development and characterization of 18 novel EST-SSRs from the Western Flower Thrips, Frankliniella occidentalis (Pergande). Int. J. Mol. Sci. 2012, 13, 2863–2876. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, Z.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Sequence data of Artemisia annua are available from the authors. |
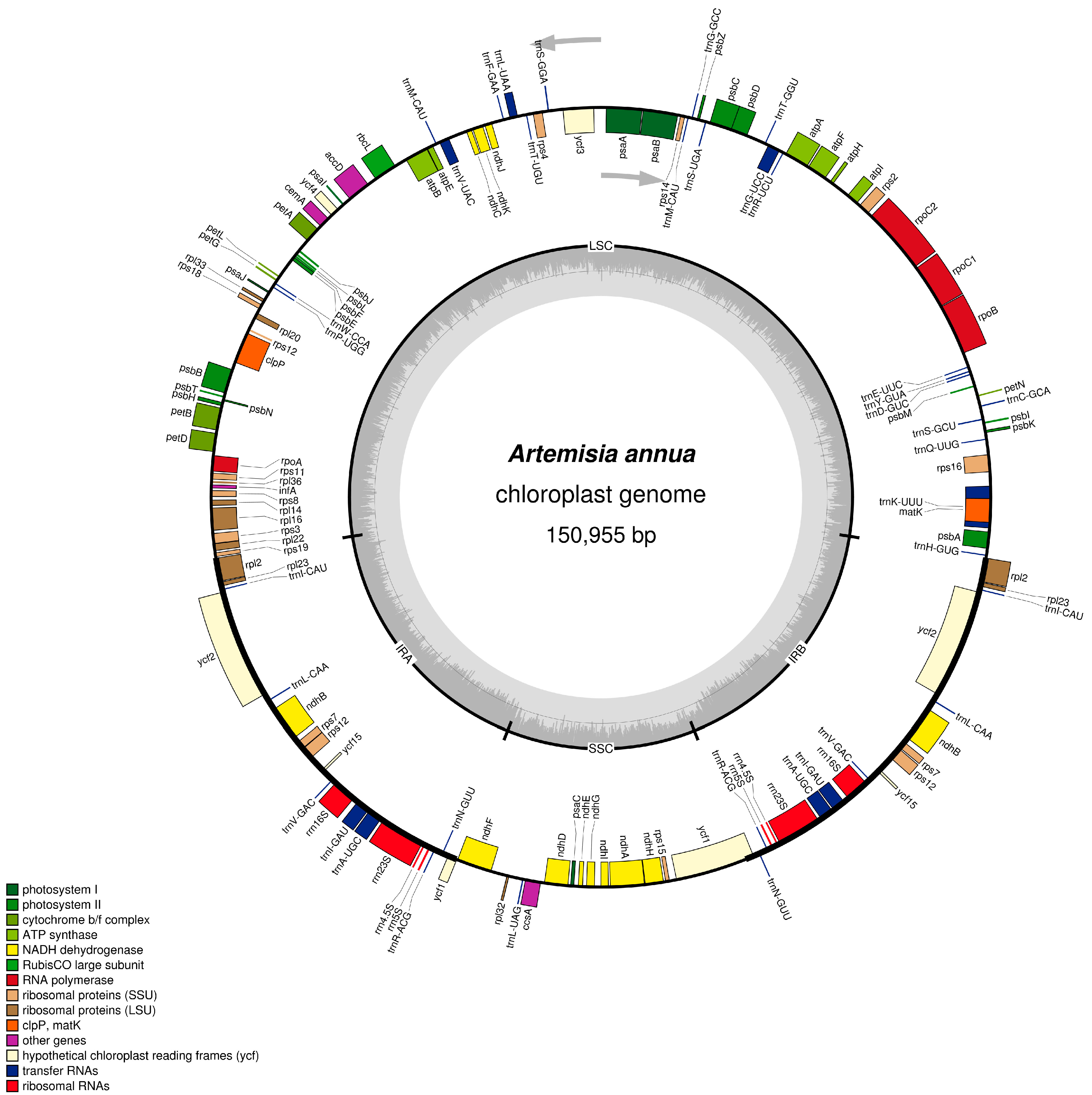
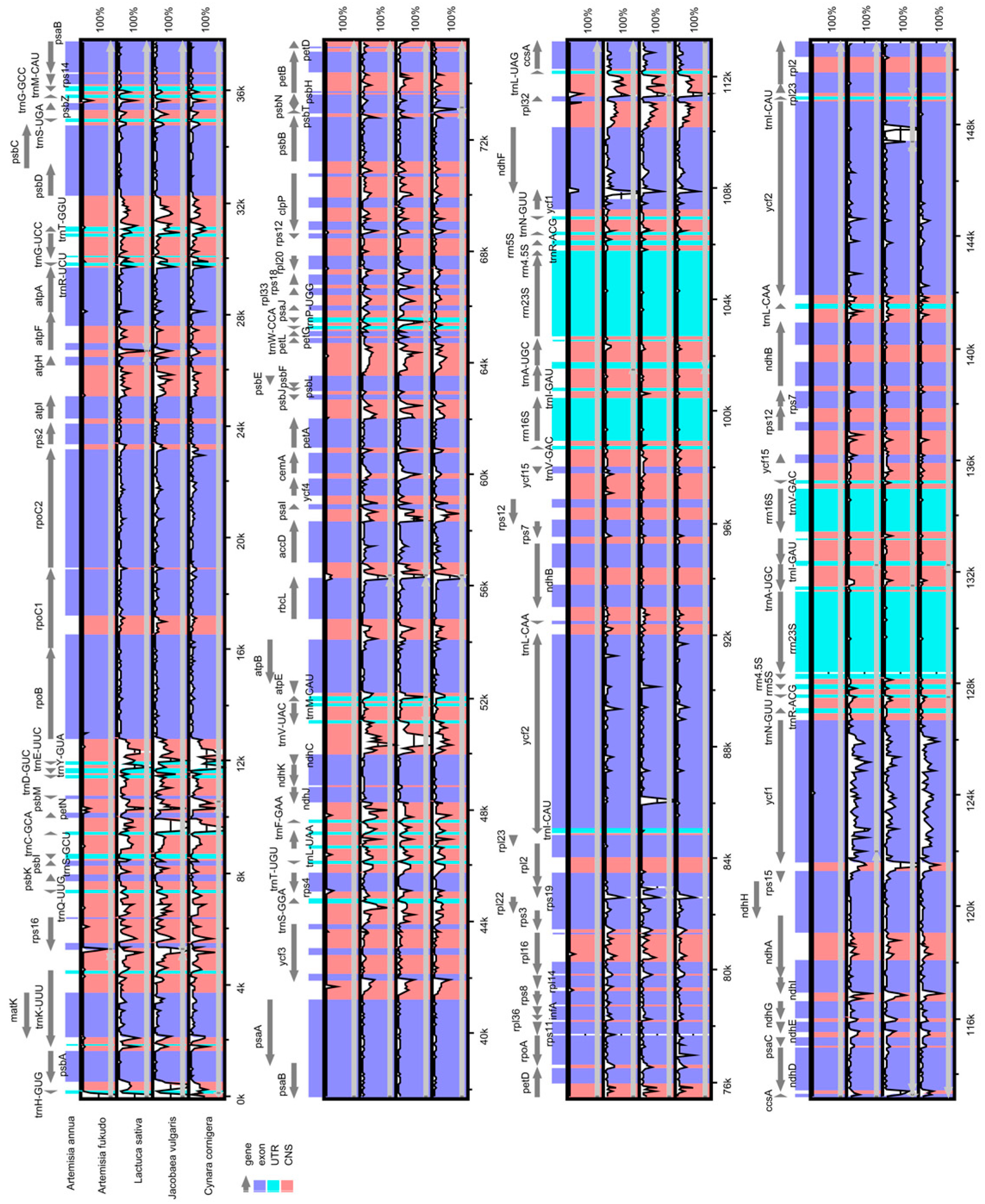

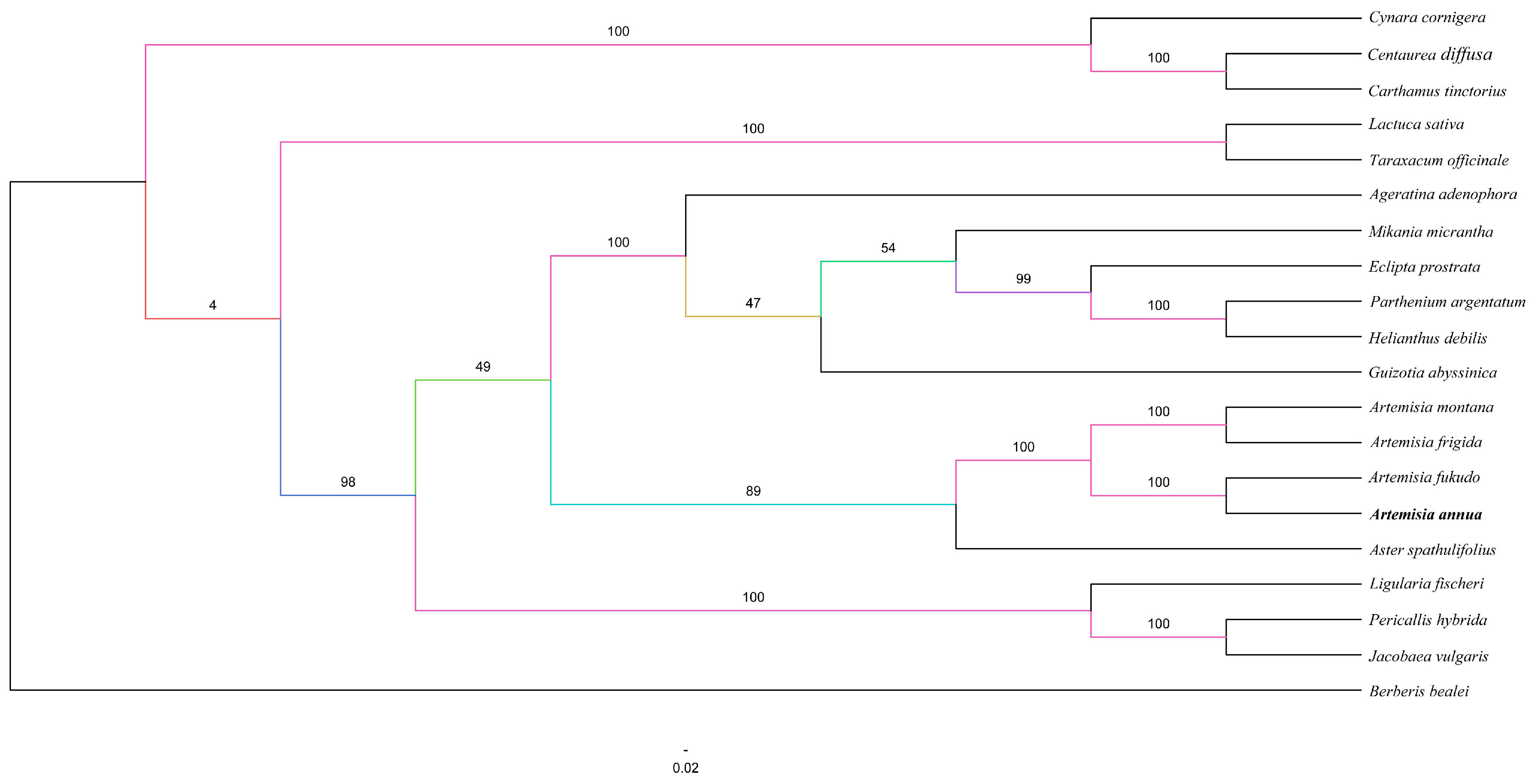
| Region | T (U) (%) | C (%) | A (%) | G (%) | Length (bp) | |
|---|---|---|---|---|---|---|
| LSC | 32.4 | 17.5 | 32.1 | 18.0 | 82,988 | |
| SSC | 34.2 | 16.1 | 35.0 | 14.7 | 18,267 | |
| IRA | 28.5 | 20.8 | 28.3 | 22.3 | 24,850 | |
| IRB | 28.3 | 22.3 | 28.5 | 20.8 | 24,850 | |
| Total | 31.3 | 18.7 | 31.2 | 18.8 | 150,955 | |
| CDS | 31.6 | 17.6 | 30.7 | 20.1 | 79,335 | |
| 1st position | 24.0 | 18.9 | 30.6 | 26.7 | 26,445 | |
| 2nd position | 33.0 | 20.2 | 29.4 | 17.7 | 26,445 | |
| 3rd position | 38.0 | 13.8 | 32.0 | 16.0 | 26,445 |
| Amino Acid | Codon | No. | RSCU | tRNA | Amino Acid | Codon | No. | RSCU | tRNA |
|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 993 | 1.32 | Tyr | UAU | 811 | 1.64 | ||
| Phe | UUC | 510 | 0.68 | trnF-GAA | Tyr | UAC | 178 | 0.36 | trnY-GUA |
| Leu | UUA | 890 | 1.87 | Stop | UAA | 52 | 1.77 | ||
| Leu | UUG | 579 | 1.22 | trnL-CAA | Stop | UAG | 21 | 0.72 | |
| Leu | CUU | 622 | 1.31 | His | CAU | 471 | 1.51 | ||
| Leu | CUC | 198 | 0.42 | His | CAC | 151 | 0.49 | trnH-GUG | |
| Leu | CUA | 368 | 0.77 | Gln | CAA | 732 | 1.52 | trnQ-UUG | |
| Leu | CUG | 196 | 0.41 | Gln | CAG | 230 | 0.48 | ||
| Ile | AUU | 1092 | 1.47 | Asn | AAU | 1017 | 1.56 | ||
| Ile | AUC | 433 | 0.58 | trnI-CAU | Asn | AAC | 287 | 0.44 | |
| Ile | AUA | 706 | 0.95 | Lys | AAA | 1042 | 1.47 | ||
| Met | AUG | 633 | 1.00 | trnM-CAU | Lys | AAG | 371 | 0.53 | |
| Val | GUU | 512 | 1.44 | Asp | GAU | 868 | 1.61 | ||
| Val | GUC | 174 | 0.49 | trnV-GAC | Asp | GAC | 213 | 0.39 | trnD-GUC |
| Val | GUA | 546 | 1.54 | Glu | GAA | 1001 | 1.50 | trnE-UUC | |
| Val | GUG | 188 | 0.53 | Glu | GAG | 337 | 0.50 | ||
| Ser | UCU | 588 | 1.74 | Cys | UGU | 202 | 1.38 | ||
| Ser | UCC | 324 | 0.96 | trnS-GGA | Cys | UGC | 91 | 0.62 | trnC-GCA |
| Ser | UCA | 417 | 1.23 | trnS-UGA | Stop | UGA | 15 | 0.51 | |
| Ser | UCG | 167 | 0.49 | Trp | UGG | 462 | 1.00 | trnW-CCA | |
| Pro | CCU | 441 | 1.58 | Arg | CGU | 350 | 1.33 | trnR-ACG | |
| Pro | CCC | 188 | 0.67 | Arg | CGC | 107 | 0.41 | ||
| Pro | CCA | 329 | 1.18 | trnP-UGG | Arg | CGA | 343 | 1.30 | |
| Pro | CCG | 159 | 0.57 | Arg | CGG | 124 | 0.47 | ||
| Thr | ACU | 535 | 1.63 | Arg | AGA | 485 | 1.84 | trnR-UCU | |
| Thr | ACC | 246 | 0.75 | trnT-GGU | Arg | AGG | 174 | 0.66 | |
| Thr | ACA | 411 | 1.25 | trnT-UGU | Ser | AGU | 410 | 1.21 | |
| Thr | ACG | 124 | 0.38 | Ser | AGC | 122 | 0.36 | trnS-GCU | |
| Ala | GCU | 617 | 1.74 | Gly | GGU | 589 | 1.32 | ||
| Ala | GCC | 228 | 0.64 | Gly | GGC | 189 | 0.42 | trnG-GCC | |
| Ala | GCA | 415 | 1.17 | Gly | GGA | 707 | 1.58 | ||
| Ala | GCG | 158 | 0.45 | Gly | GGG | 306 | 0.68 |
| Gene | Location | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) |
|---|---|---|---|---|---|---|
| trnK-UUU | LSC | 37 | 1860 | 35 | ||
| trnG-UCC | LSC | 23 | 729 | 47 | ||
| trnL-UAA | LSC | 37 | 424 | 50 | ||
| trnV-UAC | LSC | 38 | 572 | 37 | ||
| trnI-GAU | IR | 42 | 777 | 35 | ||
| trnA-UGC | IR | 38 | 812 | 35 | ||
| rps12 * | LSC | 232 | 535 | 26 | 114 | |
| rps16 | LSC | 40 | 876 | 185 | ||
| rpl16 | LSC | 9 | 1015 | 399 | ||
| rpl2 | IR | 394 | 626 | 470 | ||
| rpoC1 | LSC | 430 | 734 | 1640 | ||
| ndhA | SSC | 556 | 1064 | 539 | ||
| ndhB | IR | 777 | 670 | 756 | ||
| ycf3 | SSC | 127 | 700 | 230 | 735 | 153 |
| petB | LSC | 6 | 747 | 642 | ||
| atpF | LSC | 145 | 699 | 410 | ||
| clpP | LSC | 71 | 796 | 292 | 606 | 228 |
| ID | Repeat Start 1 | Type | Size (bp) | Repeat Start 2 | Mismatch (bp) | E-Value | Gene | Region |
|---|---|---|---|---|---|---|---|---|
| 1 | 8544 | F | 32 | 34,909 | −3 | 4.65E-05 | IGS | LSC |
| 2 | 28,063 | F | 31 | 29,661 | −3 | 1.69E-04 | IGS | LSC |
| 3 | 28,070 | F | 30 | 29,666 | −2 | 2.18E-05 | IGS | LSC |
| 4 | 38,054 | F | 32 | 40,278 | −2 | 1.55E-06 | psaB; psaA | LSC |
| 5 | 38,065 | F | 30 | 40,289 | −3 | 6.09E-04 | psaB; psaA | LSC |
| 6 | 43,070 | F | 41 | 96,883 | −1 | 1.63E-13 | ycf3 (intron); IGS | LSC; IRA |
| 7 | 43,072 | F | 39 | 118,107 | −1 | 2.48E-12 | ycf3 (intron); ndhA (intron) | LSC; SSC |
| 8 | 43,075 | F | 35 | 93,834 | −3 | 9.59E-07 | ycf3 (intron); ndhB (intron) | LSC; IRA |
| 9 | 66,346 | F | 30 | 98,046 | −2 | 2.18E-05 | IGS | LSC; IRA |
| 11 | 86,539 | F | 30 | 147,378 | −3 | 6.09E-04 | ycf2 | IRA; IRB |
| 12 | 90,121 | F | 30 | 90,157 | −1 | 5.00E-07 | ycf2 | IRA |
| 13 | 96,885 | F | 39 | 118,107 | 0 | 2.12E-14 | IGS; ndhA (intron) | IRA; SSC |
| 14 | 105,777 | F | 30 | 105,809 | −2 | 2.18E-05 | IGS | IRA |
| 15 | 128,104 | F | 30 | 128,136 | −2 | 2.18E-05 | IGS | IRB |
| 16 | 8548 | I | 30 | 44,753 | −2 | 2.18E-05 | IGS | LSC |
| 17 | 29,662 | I | 30 | 29,881 | −2 | 2.18E-05 | IGS | LSC |
| 18 | 34,911 | I | 30 | 44,755 | −1 | 5.00E-07 | IGS | LSC |
| 19 | 43,070 | I | 41 | 137,019 | −1 | 1.63E-13 | ycf3 (intron); IGS | LSC; IRB |
| 20 | 43,075 | I | 35 | 140,074 | −3 | 9.59E-07 | ycf3 (intron); ndhB (intron) | LSC; IRB |
| 21 | 66,346 | I | 30 | 135,867 | −2 | 2.18E-05 | IGS | LSC; IRB |
| 22 | 90,109 | I | 60 | 143,756 | −2 | 7.68E-23 | ycf2 | IRA; IRB |
| 23 | 90,109 | I | 42 | 143,756 | −2 | 2.57E-12 | ycf2 | IRA; IRB |
| 24 | 90,121 | I | 30 | 143,756 | −1 | 5.00E-07 | ycf2 | IRA; IRB |
| 25 | 90,124 | I | 45 | 143,756 | 0 | 5.18E-18 | ycf2 | IRA; IRB |
| 26 | 90,127 | I | 60 | 143,774 | −2 | 7.68E-23 | ycf2 | IRA; IRB |
| 27 | 90,142 | I | 45 | 143,774 | 0 | 5.18E-18 | ycf2 | IRA; IRB |
| 28 | 90,145 | I | 42 | 143,792 | −2 | 2.57E-12 | ycf2 | IRA; IRB |
| 29 | 90,157 | I | 30 | 143,792 | −1 | 5.00E-07 | ycf2 | IRA; IRB |
| 30 | 105,777 | I | 30 | 128,104 | −2 | 2.18E-05 | IGS | IRA; IRB |
| 31 | 105,809 | I | 30 | 128,136 | −2 | 2.18E-05 | IGS | IRA; IRB |
| 32 | 118,107 | I | 39 | 137,019 | 0 | 2.12E-14 | ndhA (intron); rps12 (CDS) | SSC; IRB |
| cpSSR ID | Repeat Motif | Length (bp) | Start | End | Region | Annotation |
|---|---|---|---|---|---|---|
| 1 | (A)15 | 15 | 3204 | 3218 | LSC | matK |
| 2 | (A)14 | 14 | 3708 | 3721 | LSC | |
| 3 | (A)10 | 10 | 6121 | 6130 | LSC | |
| 4 | (T)10 | 10 | 9944 | 9953 | LSC | |
| 5 | (A)10 | 10 | 13,630 | 13,639 | LSC | rpoB |
| 6 | (A)12 | 12 | 20,826 | 20,837 | LSC | rpoC2 |
| 7 | (T)10 | 10 | 23,027 | 23,036 | LSC | rpoC2 |
| 8 | (A)11 | 11 | 26,289 | 26,299 | LSC | atpH |
| 9 | (A)14 | 14 | 28,513 | 28,526 | LSC | atpA |
| 10 | (A)11 | 11 | 39,312 | 39,322 | LSC | psaA |
| 11 | (A)10 | 10 | 48,206 | 48,215 | LSC | |
| 12 | (AT)6 | 12 | 52,028 | 52,039 | LSC | |
| 13 | (T)14 | 14 | 53,085 | 53,098 | LSC | atpB |
| 14 | (A)17 | 17 | 53,306 | 53,322 | LSC | atpB |
| 15 | (A)19 | 19 | 54,902 | 54,920 | LSC | rbcL |
| 16 | (A)10 | 10 | 56,832 | 56,841 | LSC | |
| 17 | (A)14 | 14 | 57,920 | 57,933 | LSC | accD |
| 18 | (A)11 | 11 | 59,654 | 59,664 | LSC | ycf4 |
| 19 | (T)10 | 10 | 59,775 | 59,784 | LSC | ycf4 |
| 20 | (T)10 | 10 | 64,476 | 64,485 | LSC | |
| 21 | (T)10 | 10 | 64,902 | 64,911 | LSC | |
| 22 | (A)11 | 11 | 66,255 | 66,265 | LSC | |
| 23 | (T)10 | 10 | 69,525 | 69,534 | LSC | |
| 24 | (A)14 | 14 | 70,210 | 70,223 | LSC | |
| 25 | (T)10 | 10 | 71,655 | 71,664 | LSC | psbB |
| 26 | (TA)6 | 12 | 72,640 | 72,651 | LSC | psbB |
| 27 | (T)14 | 14 | 73,210 | 73,223 | LSC | psbN |
| 28 | (A)15 | 15 | 80,929 | 80,943 | LSC | |
| 29 | (T)10 | 10 | 81,209 | 81,218 | LSC | |
| 30 | (T)11 | 11 | 101,234 | 101,244 | IRA | |
| 31 | (GAA)5 | 15 | 108,039 | 108,053 | SSC | ndhF |
| 32 | (TAA)5 | 15 | 117,240 | 117,254 | SSC | ndhI |
| 33 | (T)10 | 10 | 118,903 | 118,912 | SSC | |
| 34 | (A)14 | 14 | 121,936 | 121,949 | SSC | ycf1 |
| 35 | (A)11 | 11 | 132,700 | 132,710 | IRB |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Wu, M.; Liao, B.; Liu, Z.; Bai, R.; Xiao, S.; Li, X.; Zhang, B.; Xu, J.; Chen, S. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua. Molecules 2017, 22, 1330. https://doi.org/10.3390/molecules22081330
Shen X, Wu M, Liao B, Liu Z, Bai R, Xiao S, Li X, Zhang B, Xu J, Chen S. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua. Molecules. 2017; 22(8):1330. https://doi.org/10.3390/molecules22081330
Chicago/Turabian StyleShen, Xiaofeng, Mingli Wu, Baosheng Liao, Zhixiang Liu, Rui Bai, Shuiming Xiao, Xiwen Li, Boli Zhang, Jiang Xu, and Shilin Chen. 2017. "Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua" Molecules 22, no. 8: 1330. https://doi.org/10.3390/molecules22081330
APA StyleShen, X., Wu, M., Liao, B., Liu, Z., Bai, R., Xiao, S., Li, X., Zhang, B., Xu, J., & Chen, S. (2017). Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua. Molecules, 22(8), 1330. https://doi.org/10.3390/molecules22081330




