H2S-Mediated Protein S-Sulfhydration: A Prediction for Its Formation and Regulation
Abstract
:1. Introduction
2. Potential Forming Mechanisms of Protein S-Sulfhydration
2.1. Protein S-Sulfhydration Detection
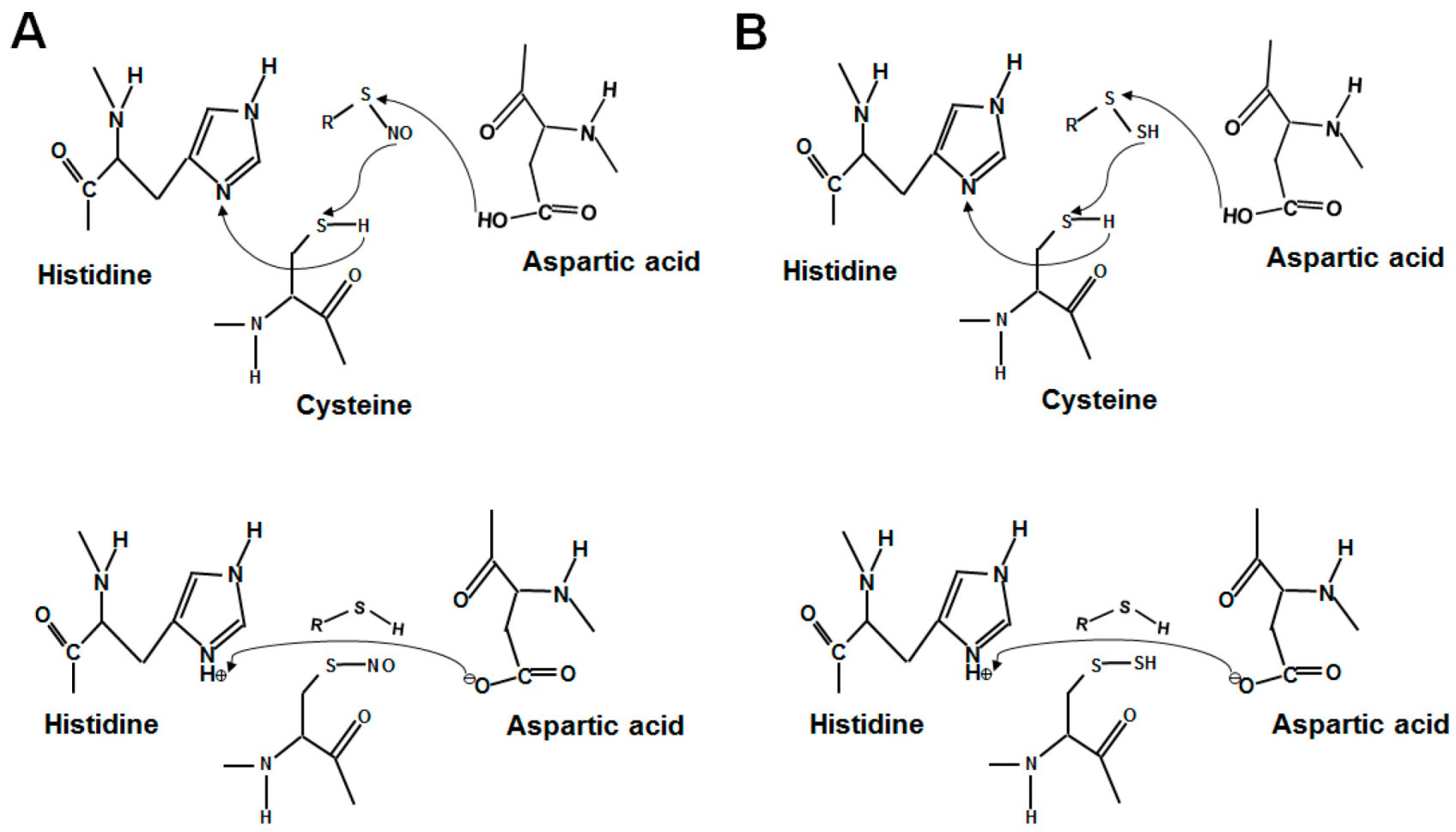
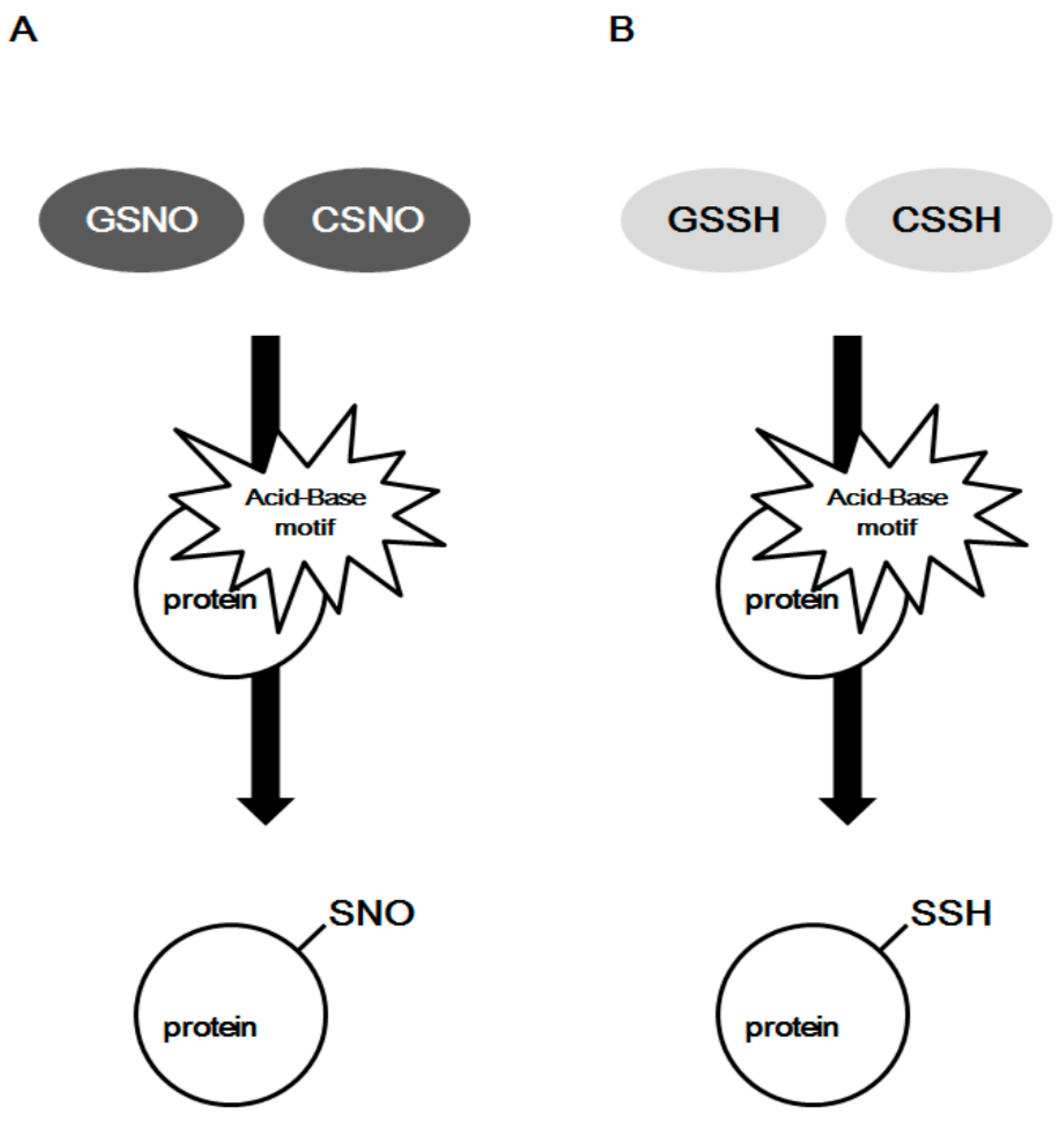
2.2. Acid–Base Motif in Protein S-Sulfhydration
2.3. Transsulfhydration via Protein–Protein Interaction
2.4. Protein S-Sulfhydration from Oxidized Cysteine (S–OH)
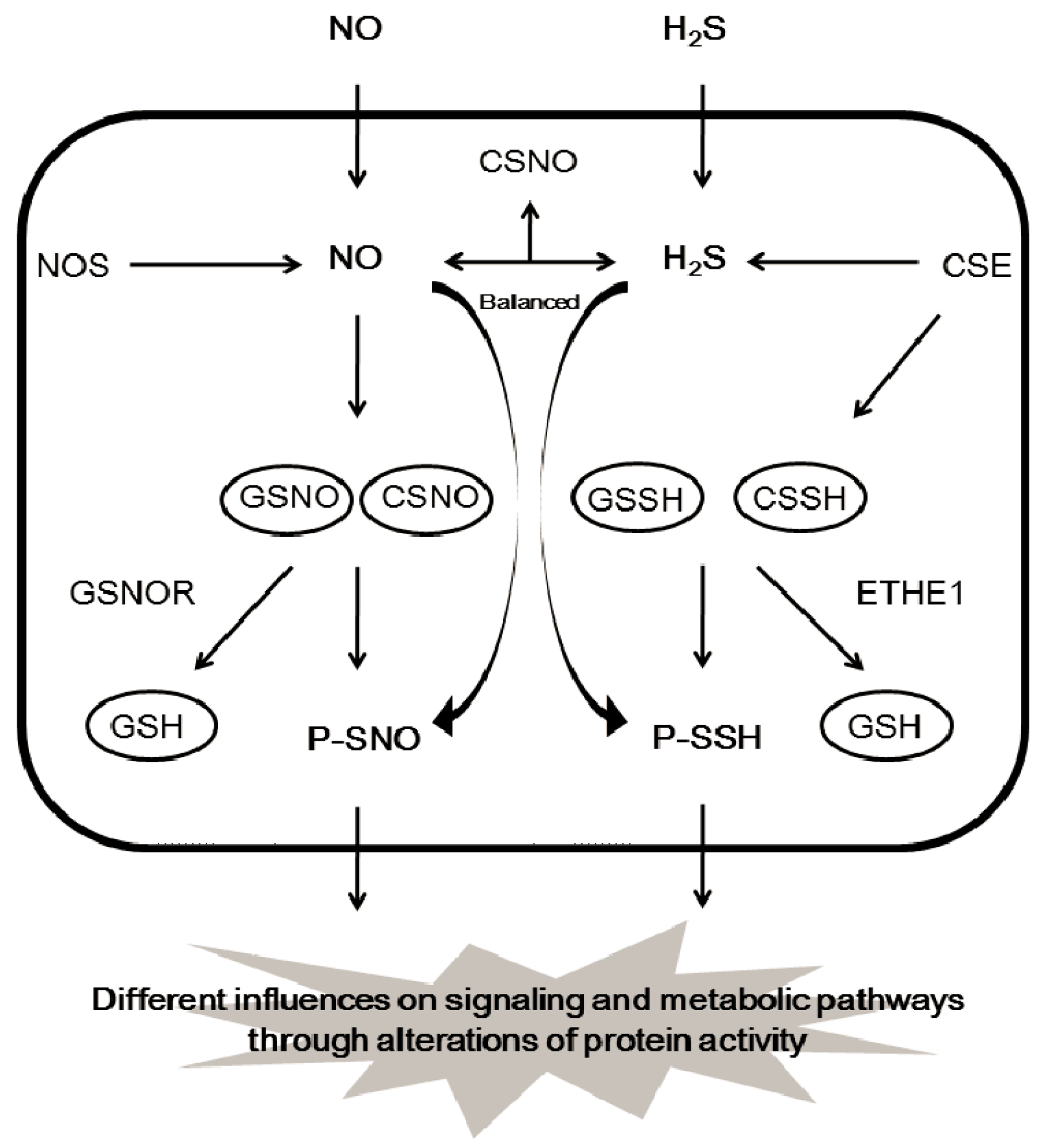
2.5. Interaction of H2S and NO on Protein Modification
3. The Donor Molecules for Protein S-Nitrosylation and S-Sulfhydration
4. Prospection
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Jaffrey, S.R.; Snyder, S.H. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE 2001, 86. [Google Scholar] [CrossRef] [PubMed]
- Jaffrey, S.R.; Erdjument-Bromage, H.; Ferris, C.D.; Tempst, P.; Snyder, S.H. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat. Cell Biol. 2001, 3, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.; Derakhshan, B.; Shi, L.; Campagne, F.; Gross, S.S. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. USA 2006, 103, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Li, B.Q.; Hu, L.L.; Niu, S.; Cai, Y.D.; Chou, K.C. Predict and analyze S-nitrosylation modification sites with the mRMR and IFS approaches. J. Proteom. 2012, 75, 1654–1665. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ding, J.; Wu, L.Y.; Chou, K.C. iSNO-PseAAC: Predict cysteine S-nitrosylation sites in proteins by incorporating position specific amino acid propensity into pseudo amino acid composition. PLoS ONE 2013, 8, e55844. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Liu, Z.; Gao, X.; Jin, C.; Wen, L.; Yao, X.; Ren, J. GPS-SNO: Computational prediction of protein S-nitrosylation sites with a modified GPS algorithm. PLoS ONE 2010, 5, e11290. [Google Scholar] [CrossRef] [PubMed]
- Ascenzi, P.; Colasanti, M.; Persichini, T.; Muolo, M.; Polticelli, F.; Venturini, G.; Bordo, D.; Bolognesi, M. Re-evaluation of amino acid sequence and structural consensus rules for cysteine-nitric oxide reactivity. Biol. Chem. 2000, 381, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Marino, S.M.; Gladyshev, V.N. Structural analysis of cysteine S-nitrosylation: A modified acid-based motif and the emerging role of trans-nitrosylation. J. Mol. Biol. 2010, 395, 844–859. [Google Scholar] [CrossRef] [PubMed]
- Stamler, J.S.; Toone, E.J.; Lipton, S.A.; Sucher, N.J. (S)NO signals: Translocation, regulation, and a consensus motif. Neuron 1997, 18, 691–696. [Google Scholar] [CrossRef]
- Mustafa, A.K.; Gadalla, M.M.; Sen, N.; Kim, S.; Mu, W.; Gazi, S.K.; Barrow, R.K.; Yang, G.; Wang, R.; Snyder, S.H. H2S signals through protein S-sulfhydration. Sci. Signal. 2009, 2, ra72. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.D.; Snyder, S.H. H2S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.D.; Snyder, S.H. Protein sulfhydration. Methods Enzymol. 2015, 555, 79–90. [Google Scholar] [PubMed]
- Paul, B.D.; Snyder, S.H. H2S: A novel gasotransmitter that signals by sulfhydration. Trends Biochem. Sci. 2015, 40, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Yang, G. Protein S-sulfhydration as a major source of H2S bioactivity. Recept. Clin. Investig. 2014, 1, e337. [Google Scholar]
- Mustafa, A.K.; Sikka, G.; Gazi, S.K.; Steppan, J.; Jung, S.M.; Bhunia, A.K.; Barodka, V.M.; Gazi, F.K.; Barrow, R.K.; Wang, R.; et al. Hydrogen sulfide as endothelium-derived hyperpolarizing factor sulfhydrates potassium channels. Circ. Res. 2011, 109, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhao, K.; Ju, Y.; Mani, S.; Cao, Q.; Puukila, S.; Khaper, N.; Wu, L.; Wang, R. Hydrogen sulfide protects against cellular senescence via S-sulfhydration of Keap1 and activation of Nrf2. Antioxid. Redox Signal. 2013, 18, 1906–1919. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Ju, Y.; Li, S.; Al Tanny, Z.; Wang, R.; Yang, G. S-sulfhydration of MEK1 leads to PARP-1 activation and DNA damage repair. EMBO Rep. 2014, 15, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Altaany, Z.; Ju, Y.; Yang, G.; Wang, R. The coordination of S-sulfhydration, S-nitrosylation, and phosphorylation of endothelial nitric oxide synthase by hydrogen sulfide. Sci. Signal. 2014, 7, ra87. [Google Scholar] [CrossRef] [PubMed]
- Pieragostino, D.; Del Boccio, P.; Di Ioia, M.; Pieroni, L.; Greco, V.; De Luca, G.; D’Aguanno, S.; Rossi, C.; Franciotta, D.; Centonze, D.; et al. Oxidative modifications of cerebral transthyretin are associated with multiple sclerosis. Proteomics 2013, 13, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.Z.; Shi, M.M.; Xie, L.; Wu, Z.Y.; Li, G.; Hua, F.; Bian, J.S. Sulfhydration of p66Shc at cysteine59 mediates the anti-oxidant effect of Hydrogen Sulfide. Antioxid. Redox Signal. 2014, 21, 2531–2542. [Google Scholar] [CrossRef] [PubMed]
- Vandiver, M.S.; Paul, B.D.; Xu, R.; Karuppagounder, S.; Rao, F.; Snowman, A.M.; Ko, H.S.; Lee, Y.I.; Dawson, V.L.; Dawson, T.M.; et al. Sulfhydration mediates neuroprotective actions of parkin. Nat. Commun. 2013, 4, 1626. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, N.; Fu, C.; Pappin, D.J.; Tonks, N.K. H2S-induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Sci. Signal. 2011, 4, ra86. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P. Mechanistic chemical perspective of hydrogen sulfide signaling. Methods Enzymol. 2015, 554, 3–29. [Google Scholar] [PubMed]
- Li, H.; Wan, A.; Xu, G.; Ye, D. Small changes huge impact: The role of thioredoxin 1 in the regulation of apoptosis by S-nitrosylation. Acta Biochim. Biophys. Sin. 2013, 45, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Filipovic, M.R. Persulfidation (S-sulfhydration) and H2S. Handb. Exp. Pharmacol. 2015, 230, 29–59. [Google Scholar] [PubMed]
- Ju, Y.; Fu, M.; Wu, L.; Yang, G. Strategies and tools for detection of protein S-nitrosylation and S-sulfhydration. Biochem. Anal. Biochem. 2015, 4, 224. [Google Scholar]
- Zhang, D.; Macinkovic, I.; Devarie-Baez, N.O.; Pan, J.; Park, C.M.; Carroll, K.S.; Filipovic, M.R.; Xian, M. Detection of protein S-sulfhydration by a tag-switch technique. Angew. Chem. Int. Ed. Engl. 2014, 53, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Wedmann, R.; Onderka, C.; Wei, S.; Szijártó, I.A.; Miljkovic, J.L.; Mitrovic, A.; Lange, M.; Savitsky, S.; Yadav, P.K.; Torregrossa, R.; et al. Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem. Sci. 2016, 7, 3414–3426. [Google Scholar] [CrossRef] [PubMed]
- Sen, N. Functional and molecular insights of hydrogen sulfide signaling and protein sulfhydration. J. Mol. Biol. 2017, 429, 543–561. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Kasamatsu, S.; Matsunaga, T.; Akashi, S.; Ono, K.; Nishimura, A.; Morita, M.; Abdul, H.H.; Fujii, S.; Kitamura, H.; et al. Protein polysulfidation-dependent persulfide dioxygenase activity of ethylmalonic encephalopathy protein 1. Biochem. Biophys. Res. Commun. 2016, 480, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Dóka, É.; Pader, I.; Bíró, A.; Johansson, K.; Cheng, Q.; Ballagó, K.; Prigge, J.R.; Pastor-Flores, D.; Dick, T.P.; Schmidt, E.E.; et al. A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci. Adv. 2016, 2, e1500968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Irie, Y.; Keung, W.M.; Maret, W. S-nitrosothiols react preferentially with zinc thiolate clusters of metallothionein III through transnitrosation. Biochemistry 2002, 41, 8360–8367. [Google Scholar] [CrossRef] [PubMed]
- Benhar, M.; Forrester, M.T.; Stamler, J.S. Protein denitrosylation: Enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Foster, M.W.; Hess, D.T.; Stamler, J.S. Protein S-nitrosylation in health and disease: A current perspective. Trends Mol. Med. 2009, 15, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Sayed, N.; Kim, D.D.; Fioramonti, X.; Iwahashi, T.; Durán, W.N.; Beuve, A. Nitroglycerin-induced S-nitrosylation and desensitization of soluble guanylyl cyclase contribute to nitrate tolerance. Circ. Res. 2008, 103, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Fioramonti, X.; Deak, A.; Deshpande, S.; Carneiro, L.; Zhou, C.; Sayed, N.; Orban, B.; Berlin, J.R.; Pénicaud, L.; Leloup, C.; et al. Hypothalamic S-nitrosylation contributes to the counter-regulatory response impairment following recurrent hypoglycemia. PLoS ONE 2013, 8, e68709. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.X.; Li, S.; Whorton, A.R. Redox regulation of PTEN by S-nitrosothiols. Mol. Pharmacol. 2005, 68, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Cortese-Krott, M.M.; Kuhnle, G.G.; Dyson, A.; Fernandez, B.O.; Grman, M.; DuMond, J.F.; Barrow, M.P.; McLeod, G.; Nakagawa, H.; Ondrias, K.; et al. Key bioactive reaction products of the NO/H2S interaction are S/N-hybrid species, polysulfides, and nitroxyl. Proc. Natl. Acad. Sci. USA 2015, 112, 4651–4660. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Kavalier, A.; Lukyanov, E.; Gross, S.S. S-sulfhydration/desulfhydration and S-nitrosylation/denitrosylation: A common paradigm for gasotransmitter signaling by H2S and NO. Methods 2013, 62, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Ida, T.; Sawa, T.; Ihara, H.; Tsuchiya, Y.; Watanabe, Y.; Kumagai, Y.; Suematsu, M.; Motohashi, H.; Fujii, S.; Matsunaga, T.; et al. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 7606–7611. [Google Scholar] [CrossRef] [PubMed]
- Kabil, O.; Banerjee, R. Characterization of patient mutations in human persulfide dioxygenase (ETHE1) involved in H2S catabolism. J. Biol. Chem. 2012, 287, 44561–44567. [Google Scholar] [CrossRef] [PubMed]
- Greiner, R.; Pálinkás, Z.; Bäsell, K.; Becher, D.; Antelmann, H.; Nagy, P.; Dick, T.P. Polysulfides link H2S to protein thiol oxidation. Antioxid. Redox Signal. 2013, 19, 1749–1765. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Untereiner, A.; Wu, L.; Yang, G. H2S-induced S-sulfhydration of pyruvate carboxylase contributes to gluconeogenesis in liver cells. Biochim. Biophys. Acta 2015, 1850, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Kasamatsu, S.; Nishimura, A.; Morita, M.; Matsunaga, T.; Abdul, H.H.; Akaike, T. Redox signaling regulated by cysteine persulfide and protein polysulfidation. Molecules 2016, 21, 1721. [Google Scholar] [CrossRef] [PubMed]
- Vasas, A.; Dóka, É.; Fábián, I.; Nagy, P. Kinetic and thermodynamic studies on the disulfide-bond reducing potential of hydrogen sulfide. Nitric Oxide 2015, 46, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Mikami, Y.; Shibuya, N.; Kimura, Y.; Nagahara, N.; Ogasawara, Y.; Kimura, H. Thioredoxin and dihydrolipoic acid are required for 3-mercaptopyruvate sulfurtransferase to produce hydrogen sulfide. Biochem. J. 2011, 439, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Haendeler, J.; Hoffmann, J.; Tischler, V.; Berk, B.C.; Zeiher, A.M.; Dimmeler, S. Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nat. Cell Biol. 2002, 4, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, Z.T.; Sha, Y.; Foster, M.W.; Foster, W.M.; Forrester, M.T.; Marshall, H.E. Thioredoxin-mediated denitrosylation regulates cytokine-induced nuclear factor κB (NF-κB) activation. J. Biol. Chem. 2014, 289, 3066–3072. [Google Scholar] [CrossRef] [PubMed]
- Benhar, M.; Forrester, M.T.; Hess, D.T.; Stamler, J.S. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 2008, 320, 1050–1054. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Parrott, A.M.; Liu, T.; Beuve, A.; Li, H. Functional proteomics approaches for the identification of transnitrosylase and denitrosylase targets. Methods 2013, 62, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Liu, T.; Chen, W.; Oka, S.; Fu, C.; Jain, M.R.; Parrott, A.M.; Baykal, A.T.; Sadoshima, J.; Li, H. Redox regulatory mechanism of transnitrosylation by thioredoxin. Mol. Cell. Proteom. 2010, 9, 2262–2275. [Google Scholar] [CrossRef] [PubMed]
- Sumbayev, V.V. S-nitrosylation of thioredoxin mediates activation of apoptosis signal-regulating kinase 1. Arch. Biochem. Biophys. 2003, 415, 133–136. [Google Scholar] [CrossRef]
- Mitchell, D.A.; Morton, S.U.; Fernhoff, N.B.; Marletta, M.A. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc. Natl. Acad. Sci. USA 2007, 104, 11609–11614. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, M.D.; Sen, N.; Hara, M.R.; Juluri, K.R.; Nguyen, J.V.; Snowman, A.M.; Law, L.; Hester, L.D.; Snyder, S.H. GAPDH mediates nitrosylation of nuclear proteins. Nat. Cell Biol. 2010, 12, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Pawloski, J.R.; Hess, D.T.; Stamler, J.S. Export by red blood cells of nitric oxide bioactivity. Nature 2001, 409, 622–626. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Kim, C.K.; Lee, K.H.; Ahn, J.Y. S-nitrosylation of B23/nucleophosmin by GAPDH protects cells from the SIAH1-GAPDH death cascade. J. Cell Biol. 2012, 199, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Wu, L.; Yang, G. Thioredoxin 1 regulation of protein S-desulfhydration. Biochem. Biophys. Rep. 2016, 5, 27–34. [Google Scholar] [CrossRef]
- Murray, C.I.; Van Eyk, J.E. Chasing cysteine oxidative modifications: proteomic tools for characterizing cysteine redox status. Circ. Cardiovasc. Genet. 2012, 5, 591. [Google Scholar] [CrossRef] [PubMed]
- Lo Conte, M.; Carroll, K.S. The redox biochemistry of protein sulfenylation and sulfinylation. J. Biol. Chem. 2013, 288, 26480–26488. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Kirber, M.T.; Xiao, H.; Yang, Y.; Keaney, J.F. Regulation of ROS signal transduction by NADPH oxidase 4 localization. J. Cell Biol. 2008, 181, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, L.; Bianco, C.L.; Toscano, J.P.; Lin, J.; Akaike, T.; Fukuto, J.M. Chemical biology of hydropersulfides and related species: Possible roles in cellular protection and redox signaling. Antioxid. Redox Signal. 2017. Epub ahead of print. [Google Scholar]
- Bianco, C.L.; Fukuto, J.M. Examining the reaction of NO and H2S and the possible cross-talk between the two signaling pathways. Proc. Natl. Acad. Sci. USA 2015, 112, 10573–10574. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Kevil, C.G. Nitric oxide and hydrogen sulfide regulation of ischemic vascular remodeling. Microcirculation 2016, 23, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Kolluru, G.K.; Yuan, S.; Shen, X.; Kevil, C.G. H2S regulation of nitric oxide metabolism. Methods Enzymol. 2015, 554, 271–297. [Google Scholar] [PubMed]
- Filipovic, M.R.; Miljkovic, J.L.; Nauser, T.; Royzen, M.; Klos, K.; Shubina, T.; Koppenol, W.H.; Lippard, S.J.; Ivanović-Burmazović, I. Chemical characterization of the smallest S-nitrosothiol, HSNO; cellular cross-talk of H2S and S-nitrosothiols. J. Am. Chem. Soc. 2012, 134, 12016–12027. [Google Scholar] [CrossRef] [PubMed]
- Cortese-Krott, M.M.; Fernandez, B.O.; Santos, J.L.; Mergia, E.; Grman, M.; Nagy, P.; Kelm, M.; Butler, A.; Feelisch, M. Nitrosopersulfide (SSNO(−)) accounts for sustained NO bioactivity of S-nitrosothiols following reaction with sulfide. Redox Biol. 2014, 2, 234–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulsen, C.E.; Truong, T.H.; Garcia, F.J.; Homann, A.; Gupta, V.; Leonard, S.E.; Carroll, K.S. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol. 2012, 8, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Lamotte, O.; Bertoldo, J.B.; Besson-Bard, A.; Rosnoblet, C.; Aimé, S.; Hichami, S.; Terenzi, H.; Wendehenne, D. Protein S-nitrosylation: Specificity and identification strategies in plants. Front. Chem. 2014, 2, 114. [Google Scholar] [CrossRef] [PubMed]
- Hess, D.T.; Matsumoto, A.; Kim, S.O.; Marshall, H.E.; Stamler, J.S. Protein S-nitrosylation: Purview and parameters. Nat. Rev. Mol. Cell Biol. 2005, 6, 150–166. [Google Scholar] [CrossRef] [PubMed]
- Hill, B.G.; Dranka, B.P.; Bailey, S.M.; Lancaster, J.R.; Darley-Usmar, V.M. What part of NO don’t you understand? Some answers to the cardinal questions in nitric oxide biology. J. Biol. Chem. 2010, 285, 19699–19704. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.C.; Marletta, M.A. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr. Opin. Chem. Biol. 2012, 16, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Saund, S.S.; Sosa, V.; Henriquez, S.; Nguyen, Q.N.; Bianco, C.L.; Soeda, S.; Millikin, R.; White, C.; Le, H.; Ono, K.; et al. The chemical biology of hydropersulfides (RSSH): Chemical stability, reactivity and redox roles. Arch. Biochem. Biophys. 2015, 15, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.S.; Pluth, M.D. Reactions of isolated persulfides provide insights into the interplay between H2S and persulfide reactivity. Free Radic. Biol. Med. 2015, 89, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Libiad, M.; Yadav, P.K.; Vitvitsky, V.; Martinov, M.; Banerjee, R. Organization of the human mitochondrial hydrogen sulfide oxidation pathway. J. Biol. Chem. 2014, 289, 30901–30910. [Google Scholar] [CrossRef] [PubMed]


| Regulatory Protein | Target Protein | Reference |
|---|---|---|
| Trx1 | Alpha enolase | [51] |
| Heat shock cognate 71 kDa protein | [51] | |
| Peroxiredoxin-1 | [51] | |
| Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) | Deacetylating enzyme sirtuin-1 | [54] |
| Histone deacetylase-2 | [54] | |
| DNA-activated protein kinase | [54] | |
| B23/nucleophosmin | [56] | |
| Haemoglobin | Anion exchanger AE1 | [55] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ju, Y.; Fu, M.; Stokes, E.; Wu, L.; Yang, G. H2S-Mediated Protein S-Sulfhydration: A Prediction for Its Formation and Regulation. Molecules 2017, 22, 1334. https://doi.org/10.3390/molecules22081334
Ju Y, Fu M, Stokes E, Wu L, Yang G. H2S-Mediated Protein S-Sulfhydration: A Prediction for Its Formation and Regulation. Molecules. 2017; 22(8):1334. https://doi.org/10.3390/molecules22081334
Chicago/Turabian StyleJu, Youngjun, Ming Fu, Eric Stokes, Lingyun Wu, and Guangdong Yang. 2017. "H2S-Mediated Protein S-Sulfhydration: A Prediction for Its Formation and Regulation" Molecules 22, no. 8: 1334. https://doi.org/10.3390/molecules22081334






