Computational Studies on Acetylcholinesterases
Abstract
:1. Introduction
2. Results
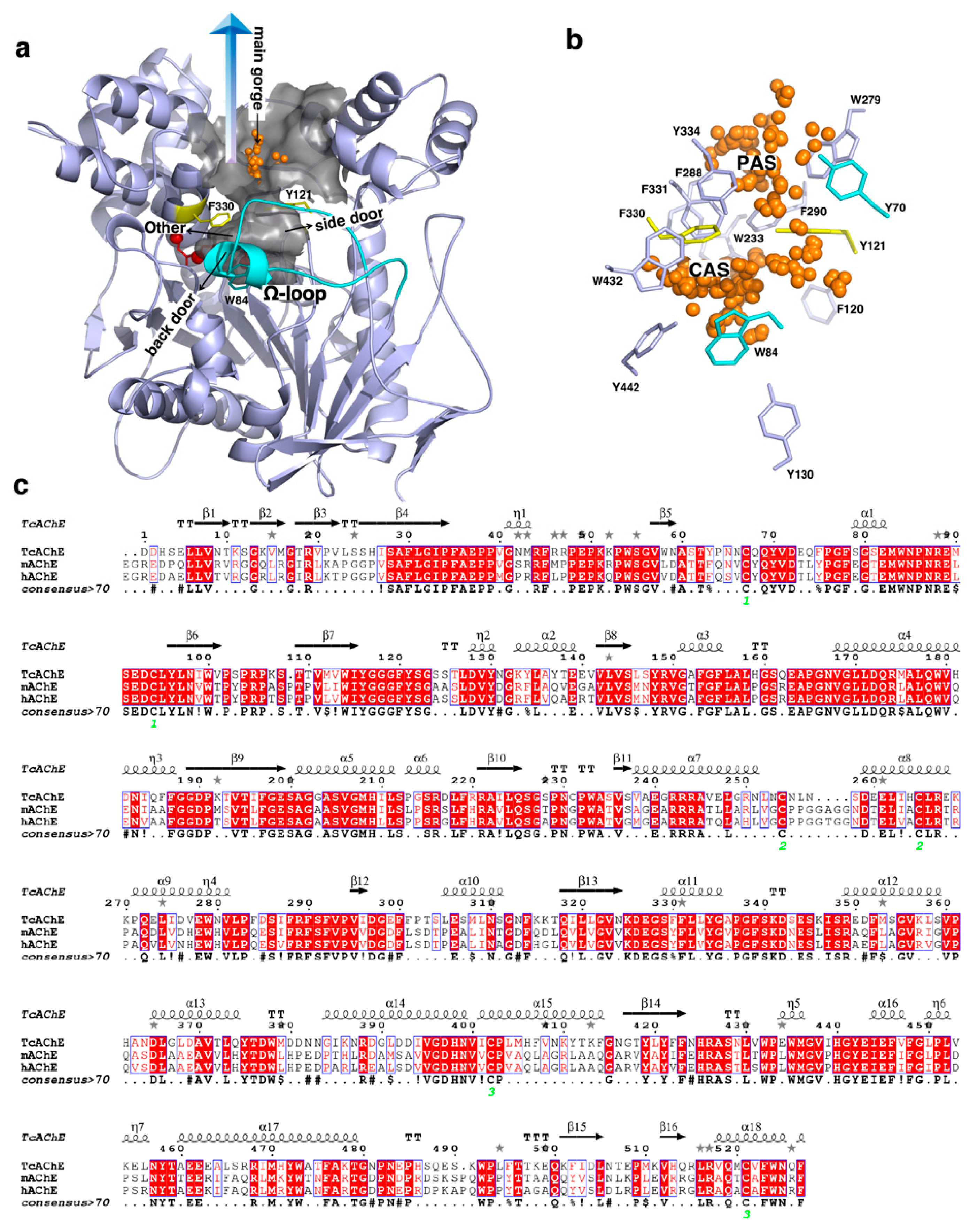
2.1. Features of the Active-Site Gorge
2.1.1. Gating of the Active-Site Gorge
2.1.2. The Role of the Electrostatic Potential in Binding of Cationic Ligands
2.1.3. Flexibility of the Aromatic Residues inside the Gorge
2.1.4. Flexibility of the Ω-Loop
2.1.5. Properties of Water Molecules inside and outside the Active-Site Gorge
2.2. Ligand Trafficking Pathways: Back Door and Side Door
2.3. Ligand Trafficking within AChE
2.3.1. ACh, Choline and their Analogues
2.3.2. HupA
2.3.3. E2020
2.3.4. Fasculin-2
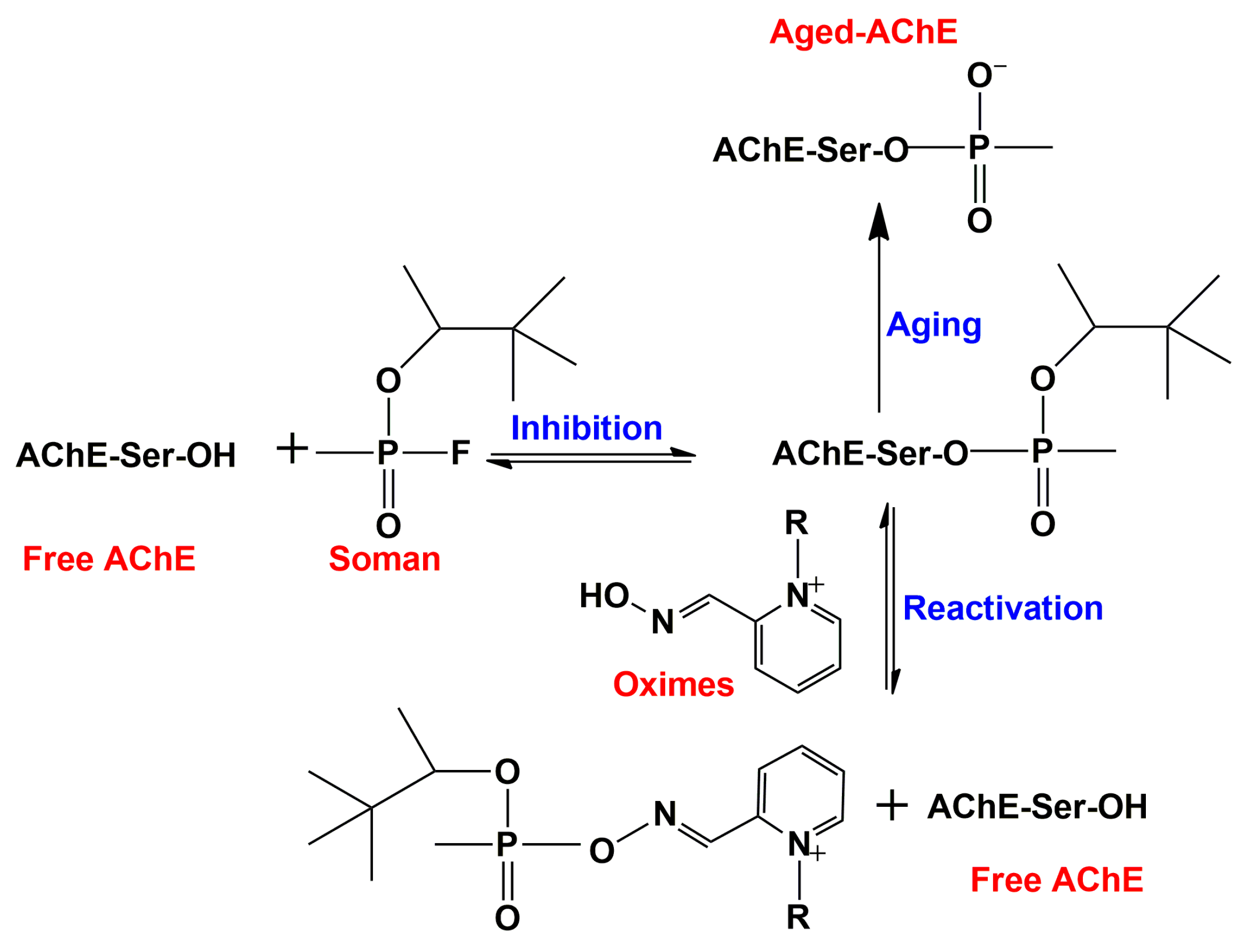
2.3.5. Interaction of Quaternary Oximes with Conjugates of AChE with Organophosphate Nerve Agents
2.3.6. syn-TZ2PA6
2.4. Dynamics of Oligomeric AChE
2.5. Catalytic Reaction Mechanism
3. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Kumar, A.; Nisha, C.M.; Silakari, C.; Sharma, I.; Anusha, K.; Gupta, N.; Nair, P.; Tripathi, T.; Kumar, A. Current and novel therapeutic molecules and targets in alzheimer’s disease. J. Formos. Med. Assoc. 2016, 115, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Rosenberry, T.L. Acetylcholinesterase. Adv. Enzymol. Relat. Areas Mol. Biol. 1975, 43, 103–218. [Google Scholar] [PubMed]
- Silman, I.; Sussman, J.L. Acetylcholinesterase: ‘Classical’ and ‘non-classical’ functions and pharmacology. Curr. Opin. Phamacol. 2005, 5, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Pohanka, M. Acetylcholinesterase inhibitors: A patent view (2008-present). Expert Opin. Ther. Pat. 2012, 22, 871–886. [Google Scholar] [CrossRef] [PubMed]
- McHardy, S.F.; Wang, H.L.; McCowen, S.V.; Valdez, M.C. Recent advances in acetylcholinesterase inhibitors and reactivators: An update on the patent literature (2012–2015). Expert Opin. Ther. Pat. 2017, 27, 455–476. [Google Scholar] [CrossRef] [PubMed]
- Sussman, J.L.; Harel, M.; Frolow, F.; Oefner, C.; Goldman, A.; Toker, L.; Silman, I. Atomic structure of acetylcholinesterase from Torpedo californica: A prototypic acetylcholine-binding protein. Science 1991, 253, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Bourne, Y.; Taylor, P.; Marchot, P. Acetylcholinesterase inhibition by fasciculin: Crystal structure of the complex. Cell 1995, 83, 503–512. [Google Scholar] [CrossRef]
- Kryger, G.; Harel, M.; Giles, K.; Toker, L.; Velan, B.; Lazar, A.; Kronman, C.; Barak, D.; Ariel, N.; Shafferman, A.; et al. Structures of recombinant native and E202Q mutant human acetylcholinesterase complexed with the snake-venom toxin fasciculin-ii. Acta Crystallogr. D Biol. Crystallogr. 2000, 56, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Colletier, J.P.; Fournier, D.; Greenblatt, H.M.; Stojan, J.; Sussman, J.L.; Zaccai, G.; Silman, I.; Weik, M. Structural insights into substrate traffic and inhibition in acetylcholinesterase. EMBO J. 2006, 25, 2746–2756. [Google Scholar] [CrossRef] [PubMed]
- Kryger, G.; Silman, I.; Sussman, J.L. Structure of acetylcholinesterase complexed with E2020 (Aricept):Implications for the design of new anti-alzheimer drugs. Structure 1999, 7, 297–307. [Google Scholar] [CrossRef]
- Ripoll, D.R.; Faerman, C.H.; Axelsen, P.H.; Silman, I.; Sussman, J.L. An electrostatic mechanism for substrate guidance down the aromatic gorge of acetylcholinesterase. Proc. Natl. Acad. Sci. USA 1993, 90, 5128–5132. [Google Scholar] [CrossRef] [PubMed]
- McCammon, J.A.; Gelin, B.R.; Karplus, M. Dynamics of folded proteins. Nature 1977, 267, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Tai, K.; Henchman, R.H.; McCammon, J.A. Molecular dynamics of acetylcholinesterase. Acc. Chem. Res. 2002, 35, 332–340. [Google Scholar] [CrossRef] [PubMed]
- The Pymol Molecular Graphics System; Version 1.8; Schrödinger LLC: New York, NY, USA, 2015.
- Le Guilloux, V.; Schmidtke, P.; Tuffery, P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinform. 2009, 10, 168. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWiliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal w and clustal x version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new endscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Wlodek, S.T.; Clark, T.W.; Scott, L.R.; McCammon, J.A. Molecular dynamics of acetylcholinesterase dimer complexed with tacrine. J. Am. Chem. Soc. 1997, 119, 9513–9522. [Google Scholar] [CrossRef]
- Zhou, H.X.; Wlodek, S.T.; McCammon, J.A. Conformation gating as a mechanism for enzyme specificity. Proc. Natl. Acad. Sci. USA 1998, 95, 9280–9283. [Google Scholar] [CrossRef] [PubMed]
- Tai, K.; Shen, T.; Börjesson, U.; Philippopoulos, M.; McCammon, J.A. Analysis of a 10-ns molecular dynamics simulation of mouse acetylcholinesterase. Biophys. J. 2001, 81, 715–724. [Google Scholar] [CrossRef]
- Cheng, S.; Song, W.; Yuan, X.; Xu, Y. Gorge motions of acetylcholinesterase revealed by microsecond molecular dynamics simulations. Sci. Rep. 2017, 7, 3219. [Google Scholar] [CrossRef] [PubMed]
- Antosiewicz, J.; Gilson, M.K.; Lee, I.H.; McCammon, J.A. Acetylcholinesterase: Diffusional encounter rate constants for dumbbell models of ligand. Biophys. J. 1995, 68, 62–68. [Google Scholar] [CrossRef]
- Antosiewicz, J.; McCammon, J.A.; Wodak, S.J.; Gilson, M.K. Simulation of charge-mutant acetylcholinesterases. Biochemistry 1995, 43, 4211–4219. [Google Scholar] [CrossRef]
- Antosiewicz, J.; Wlodek, S.T.; McCammon, J.A. Acetylcholinesterase: Role of the enzyme’s charge distribution in steering charged ligands towards the active site. Biopolymers 1996, 39, 85–94. [Google Scholar] [CrossRef]
- Wade, R.C. Brownian dynamics simulations of enzyme-substrate encounter. Biochem. Soc. Trans. 1996, 24, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Qian, H. From discrete protein kinetics to continuous Brownian dynamics: A new perspective. Protein Sci. 2002, 11, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Shafferman, A.; Ordentlich, A.; Barak, D.; Kronman, C.; Ber, R.; Bino, T.; Ariel, N.; Osman, R.; Velan, B. Electrostatic attraction by surface charge does not contribute to the catalytic efficiency of acetylcholinesterase. EMBO J. 1994, 13, 3448–3455. [Google Scholar] [PubMed]
- Zhou, H.X.; Briggs, J.M.; McCammon, J.A. A 240-fold electrostatic rate-enhancement for acetylcholinesterase-substrate binding can be predicted by the potential within the active site. J. Am. Chem. Soc. 1996, 118, 13069–13070. [Google Scholar] [CrossRef]
- Radić, Z.; Kirchhoff, P.D.; Quinn, D.M.; McCammon, J.A.; Taylor, P. Electrostatic influence on the kinetics of ligand binding to acetylcholinesterase. Distinctions between active center ligands and fasciculin. J. Biol. Chem. 1997, 272, 23265–23277. [Google Scholar] [CrossRef] [PubMed]
- Tara, S.; Elcock, A.H.; Kirchhoff, P.D.; Briggs, J.M.; Radić, Z.; Taylor, P.; McCammon, J.A. Rapid binding of a cationic active site inhibitor to wild type and mutant mouse acetylcholinesterase: Brownian dynamics simulation including diffusion in the active site gorge. Biopolymers 1998, 46, 465–474. [Google Scholar] [CrossRef]
- Elcock, A.H.; Gabdouline, R.R.; Wade, R.C.; McCammon, J.A. Computer simulation of protein-protein association kinetics: Acetylcholinesterase-fasciculin. J. Mol. Biol. 1999, 291, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Colletier, J.P.; Weik, M.; Jiang, H.; Moult, J.; Silman, I.; Sussman, J.L. Flexibility of aromatic residues in the active-site gorge of acetylcholinesterase: X-ray versus molecular dynamics. Biophys. J. 2008, 95, 2500–2511. [Google Scholar] [CrossRef] [PubMed]
- Harel, M.; Sussman, J.L.; Krejci, E.; Bon, S.; Chanal, P.; Massoulié, J.; Silman, I. Conversion of acetylcholinesterase to butyrylcholinesterase: Modeling and mutagenesis. Proc. Natl. Acad. Sci. USA 1992, 89, 10827–10831. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Pang, Y.P.; Muzka, J.L.; Zhan, C.G. Active site gating and substrate specificity of butyrylcholinesterase and acetylcholinesterase: Insights from molecular dynamics simulations. J. Phys. Chem. B 2011, 115, 8797–8805. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, D.; Ordentlich, A.; Barak, D.; Ariel, N.; Kronman, C.; Velan, B.; Shafferman, A. Does “butyrylization” of acetylcholinesterase through substitution of the six divergent aromatic amino acids in the active center gorge generate an enzyme mimic of butyrylcholinesterase? Biochemistry 2001, 40, 7433–7445. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Radić, Z.; Taylor, P. Inhibitors of different structure induce distinguishing conformations in the omega loop, Cys69-Cys96, of mouse acetylcholinesterase. J. Biol. Chem. 2002, 277, 43301–43308. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Tai, K.; McCammon, J.A.; Taylor, P.; Johnson, D.A. Nanosecond dynamics of the mouse acetylcholinesterase Cys69-Cys96 omega loop. J. Biol. Chem. 2003, 278, 30905–30911. [Google Scholar] [CrossRef] [PubMed]
- Bui, J.M.; Tai, K.; McCammon, J.A. Acetylcholinesterase: Enhanced fluctuations and alternative routes to the active site in the complex with fasciculin-2. J. Am. Chem. Soc. 2004, 126, 7198–7205. [Google Scholar] [CrossRef] [PubMed]
- Bui, J.M.; McCammon, J.A. Acetylcholinesterase: Pivotal roles of its long omega loop (Cys69-Cys96) in regulating substrate binding. Chem. Biol. Interact. 2005, 157–158, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, L.M.; Rodríguez Fris, J.A.; Morini, M.A.; Sierra, M.B.; Accordino, S.A.; Montes de Oca, J.M.; Pedroni, V.I.; Appignanesi, G.A. Hydration and nanocofined water: Insights from computer simulations. Subcell. Biochem. 2015, 71, 161–187. [Google Scholar] [PubMed]
- Henchman, R.H.; Tai, K.; Shen, T.; McCammon, J.A. Properties of water molecules in the active site gorge of acetylcholinesterase from computer simulation. Biophys. J. 2002, 82, 2671–2682. [Google Scholar] [CrossRef]
- Koellner, G.; Kryger, G.; Millard, C.B.; Silman, I.; Sussman, J.L.; Steiner, T. Active-site gorge and buried water molecules in crystal structures of acetylcholinesterase from Torpedo californica. J. Mol. Biol. 2000, 296, 713–735. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Shen, J.; Luo, X.; Silman, I.; Sussman, J.L.; Chen, K.; Jiang, H. How does huperzine A enter and leave the binding gorge of acetylcholinesterase? Steered molecular dynamics simulations. J. Am. Chem. Soc. 2003, 125, 11340–11349. [Google Scholar] [CrossRef] [PubMed]
- Henchman, R.H.; McCammon, J.A. Structural and dynamic properties of water around acetylcholinesterase. Protein Sci. 2002, 11, 2080–2090. [Google Scholar] [CrossRef] [PubMed]
- Fogarty, A.C.; Laage, D. Water dynamics in protein hydration shells: The molecular origins of the dynamical pertubation. J. Phys. Chem. B 2014, 118, 7715–7729. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.; Martinez, N.; Trovaslet, M.; Scannapieco, K.; Koza, M.M.; Masson, P.; Nachon, F. Dynamics of human acetylcholinesterase bound to non-covalent and covalent inhibitors shedding light on changes to the water network structure. Phys. Chem. Chem. Phys. 2016, 18, 12992–13001. [Google Scholar] [CrossRef] [PubMed]
- Millard, C.B.; Kryger, G.; Ordentlich, A.; Greenblatt, H.M.; Harel, M.; Raves, M.L.; Segall, Y.; Barak, D.; Shafferman, A.; Silman, I.; et al. Crystal structures of aged phosphonylated acetylcholinesterase: Nerve agent reaction products at the atomic level. Biochemistry 1999, 38, 7032–7039. [Google Scholar] [CrossRef] [PubMed]
- Gilson, M.K.; Straatsma, T.P.; McCammon, J.A.; Ripoll, D.R.; Faerman, C.H.; Axelsen, P.H.; Silman, I.; Sussman, J.L. Open “back door” in a molecular dynamics simulation of acetylcholinesterase. Science 1994, 263, 1276–1278. [Google Scholar] [CrossRef] [PubMed]
- Tara, S.; Straatsma, T.P.; McCammon, J.A. Mouse acetylcholinesterase unliganded and in complex with huperzine A: A comparison of molecular dynamics simulations. Biopolymers 1999, 50, 35–43. [Google Scholar] [CrossRef]
- Xu, Y.; Colletier, J.P.; Weik, M.; Qin, G.; Jiang, H.; Silman, I.; Sussman, J.L. Long route or shortcut? A molecular dynamics study of traffic of thiocholine within the active-site gorge of acetylcholinesterase. Biophys. J. 2010, 99, 4003–4011. [Google Scholar] [CrossRef] [PubMed]
- Sanson, B.; Colletier, J.P.; Xu, Y.; Lang, P.T.; Jiang, H.; Silman, I.; Sussman, J.L.; Weik, M. Backdoor opening mechanism in acetylcholinesterase based on X-ray crystallography and molecular dynamics simulations. Protein Sci. 2011, 20, 1114–1118. [Google Scholar] [CrossRef] [PubMed]
- Nachon, F.; Stojan, J.; Fournier, D. Insights into substrate and product traffic in the Drosophila melanogaster acetylcholinesterase active site gorge by enlarging a back channel. FEBS J. 2008, 275, 2659–2664. [Google Scholar] [CrossRef] [PubMed]
- Harel, M.; Kleywegt, G.J.; Ravelli, R.B.; Silman, I.; Sussman, J.L. Crystal structure of an acetylcholinesterase-fasciculin complex: Interaction of a three-fingered toxin from snake venom with its target. Structure 1995, 3, 1355–1366. [Google Scholar] [CrossRef]
- Remy, M.H.; Frobert, Y.; Grassi, J. Characterization of monoclonal antibodies that strongly inhibit Electrophorus electricus acetylcholinesterase. Eur. J. Biochem. 1995, 231, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Boume, Y.; Renault, L.; Essono, S.; Mondielli, G.; Lamourette, P.; Boquet, D.; Grassi, J.; Marchot, P. Molecular characterization of monoclonal antibodies that inhibit acetylcholinesterase by targeting the peripheral site and backdoor region. PLoS ONE 2013, 8, e77226. [Google Scholar]
- Boume, Y.; Renault, L.; Marchot, P. Crystal structure of snake venom acetylcholinesterase in complex with inhibitory antibody fragment fab410 bound at the peripheral site: Evidence for open and closed states of a back door channel. J. Biol. Chem. 2015, 290, 1522–1535. [Google Scholar]
- Tai, K.; Shen, T.; Henchman, R.H.; Boume, Y.; Marchot, P.; McCammon, J.A. Mechanism of acetylcholinesterase inhibition by fasciculin: A 5-ns molecular dynamics simulation. J. Am. Chem. Soc. 2002, 124, 6153–6161. [Google Scholar] [CrossRef] [PubMed]
- Bui, J.M.; McCammon, J.A. Protein complex formation by acetylcholinesterase and the neurotoxin fasciculin-2 appears to involve an induced-fit mechanism. Proc. Natl. Acad. Sci. USA 2006, 103, 15451–15456. [Google Scholar] [CrossRef] [PubMed]
- Bennion, B.J.; Essiz, S.G.; Lau, E.Y.; Fatterbert, J.L.; Emigh, A.; Lightstone, F.C. A wrench in the works of human acetylcholinesterase: Soman induced conformational changes revealed by molecular dynamics simulations. PLoS ONE 2015, 10, e0121092. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, H.M.; Guillou, C.; Guénard, D.; Argaman, A.; Botti, S.; Badet, B.; Thal, C.; Silman, I.; Sussman, J.L. The complex of a bivalent derivative of galanthamine with Torpedo acetylcholinesterase displays drastic deformation of the active-site gorge: Implications for structure-based drug design. J. Am. Chem. Soc. 2004, 126, 15405–15411. [Google Scholar] [CrossRef] [PubMed]
- Van Belle, D.; De Maria, L.; Lurcu, G.; Wodak, S.J. Pathways of ligand clearance in acetylcholinesterase by multiple copy sampling. J. Mol. Biol. 2000, 298, 705–726. [Google Scholar] [CrossRef] [PubMed]
- Kua, J.; Zhang, Y.; McCammon, J.A. Studying enzyme binding specificity in acetylcholinesterase using a combined molecular dynamics and multiple docking approach. J. Am. Chem. Soc. 2002, 124, 8260–8267. [Google Scholar] [CrossRef] [PubMed]
- Kua, J.; Zhang, Y.; Eslami, A.C.; Butler, J.R.; McCammon, J.A. Studying the roles of W86, E202, and Y337 in binding of acetylcholine to acetylcholinesterase using a combined molecular dynamics and multiple docking approach. Protein Sci. 2003, 12, 2675–2684. [Google Scholar] [CrossRef] [PubMed]
- Bui, J.M.; Henchman, R.H.; McCammon, J.A. The dynamics of ligand barrier crossing inside the acetylcholinesterase gorge. Biophys. J. 2003, 85, 2267–2272. [Google Scholar] [CrossRef]
- Bartel, C.; Karplus, M. Multidimensional adaptive umbrella sampling: Applications to main chain and side chain peptide conformations. J. Comput. Chem. 1997, 18, 1450–1462. [Google Scholar] [CrossRef]
- Zheng, Q.; Kyle, D.J. Multiple copy sampling: Rigid versus flexible protein. Proteins 1994, 19, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Lsralewitz, B.; Gao, M.; Schulten, K. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 2001, 11, 224–230. [Google Scholar] [CrossRef]
- Niu, C.; Xu, Y.; Xu, Y.; Luo, X.; Duan, W.; Silman, I.; Sussman, J.L.; Zhu, W.; Chen, K.; Shen, J.; et al. Dynamic mechanism of E2020 binding to acetylcholinesterase: A steered molecular dynamics simulation. J. Phys. Chem. B 2005, 109, 23730–23738. [Google Scholar] [CrossRef] [PubMed]
- Kesharwani, M.K.; Ganguly, B.; Das, A.; Bandyopadhyay, T. Differential binding of bispyridinium oxime drugs with acetylcholinesterase. Acta Pharmacol. Sin. 2010, 31, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Chandar, N.B.; Lo, R.; Ganguly, B. Quantum chemical and steered molecular dynamics studies for one pot solution to reactivate aged acetylcholinesterase with alkylator oxime. Chem. Biol. Interact. 2014, 223, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yan, H.; Tang, X.C. Progress in studies of huperzine A, a natural cholinesterase inhibitor from Chinese herbal medicine. Acta Pharmacol. Sin. 2006, 27, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Haviv, H.; Wong, D.M.; Silman, I.; Sussman, J.L. Bivalent ligands derived from huperzine A as acetylcholinesterase inhibitors. Curr. Top. Med. Chem. 2007, 7, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Raves, M.L.; Harel, M.; Pang, Y.P.; Silman, I.; Kozikowski, A.P.; Sussman, J.L. Structure of acetylcholinesterase complexed with the nootropic alkaloid, (−)-huperzine A. Nat. Struct. Biol. 1997, 4, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Ashani, Y.; Peggins, J.O., 3rd; Doctor, B.P. Mechanism of inhibition of cholinesterases by huperzine A. Biochem. Biophys. Res. Commun. 1992, 184, 719–726. [Google Scholar] [CrossRef]
- Mallender, W.D.; Szegletes, T.; Rosenberry, T.L. Acetylthiocholine binds to Asp74 at the peripheral site of human acetylcholinesterase as the first step in the catalytic pathway. Biochemistry 2000, 39, 7753–7763. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Xu, Y.; Chen, J.; Liu, Q.; Gu, J.; Wang, X.; Ma, J.; Li, H.; Onuchic, J.N.; Jiang, H. Free energy landscape for the binding process of huperzine A to acetylcholinesterase. Proc. Natl. Acad. Sci. USA 2013, 110, 4273–4278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, J.; Rudolph, M.J.; Burshteyn, F.; Cassidy, M.S.; Gary, E.N.; Love, J.; Franklin, M.C.; Height, J.J. Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J. Med. Chem. 2012, 55, 10282–10286. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Helms, V.; McCammon, J.A. Dynamical properties of fasciculin-2. Proteins 1999, 36, 447–453. [Google Scholar] [CrossRef]
- Worek, F.; Wille, T.; Koller, M.; Thiermann, H. Toxicology of organophosphorus compounds in view of an increasing terrorist threat. Arch. Toxicol. 2016, 90, 2131–2145. [Google Scholar] [CrossRef] [PubMed]
- Worek, F.; Thiermann, H.; Wille, T. Oximes in organophosphate poisoning: 60 years of hope and despair. Chem. Biol. Interact. 2016, 259, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Nachon, F.; Lockridge, O. Structural approach to the aging of phosphylated cholinesterases. Chem. Biol. Interact. 2010, 187, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Pathak, A.K.; Bandyopadhyay, T. Unbinding of fluorinated oxime drug from the AChE gorge in polarizable water: A well-tempered metadynamics study. Phys. Chem. Chem. Phys. 2017, 19, 5560–5569. [Google Scholar] [CrossRef] [PubMed]
- Lao, A.; Parrinello, M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA 2002, 99, 12562–12566. [Google Scholar] [CrossRef] [PubMed]
- Laio, A.; Gervasio, F.L. Metadynamics: A method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep. Prog. Phys. 2008, 71, 126601. [Google Scholar] [CrossRef]
- Senapati, S.; Bui, J.M.; McCammon, J.A. Induced fit in mouse acetylcholinesterase upon binding a femtomolar inhibitor: A molecular dynamics study. J. Med. Chem. 2005, 48, 8155–8162. [Google Scholar] [CrossRef] [PubMed]
- Boume, Y.; Kolb, H.C.; Radić, Z.; Sharpless, K.B.; Taylor, P.; Marchot, P. Freeze-frame inhibitor captures acetylcholinesterase in a unique conformation. Proc. Natl. Acad. Sci. USA 2004, 101, 1449–1454. [Google Scholar]
- Sussman, J.L.; Harel, M.; Frolow, F.; Varon, L.; Toker, L.; Futerman, A.H.; Silman, I. Purification and crystallization of a dimeric form of acetylcholinesterase from Torpedo californica subsequent to solubilization with phosphatidylinositol-specific phospholipase C. J. Mol. Biol. 1988, 203, 821–823. [Google Scholar] [CrossRef]
- Wlodek, S.T.; Shen, T.; McCammon, J.A. Electrostatic steering of substrate to acetylcholinesterase: Analysis of field fluctuations. Biopolymers 2000, 53, 265–271. [Google Scholar] [CrossRef]
- Massoulié, J. The origin of the molecular diversity and functional anchoring of cholinesterases. Neuro-Signals 2002, 11, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Gorfe, A.A.; Chang, C.E.; lvanov, I.; McCammon, J.A. Dynamics of the acetylcholinesterase tetramer. Biophys. J. 2008, 94, 1144–1154. [Google Scholar] [CrossRef] [PubMed]
- Gorfe, A.A.; Lu, B.; Yu, Z.; McCammon, J.A. Enzymatic activity versus structural dynamics: The case of acetylcholinesterase tetramer. Biophys. J. 2009, 97, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Barbault, F.; Maurel, F. Simulation with quantum mechanics/molecular mechanics for drug discovery. Expert Opin. Drug Discov. 2015, 10, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kua, J.; McCammon, J.A. Role of the catalytic triad and oxyanion hole in acetylcholinesterase catalysis: An ab initio QM/MM study. J. Am. Chem. Soc. 2002, 124, 10572–10577. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, S.; Zhang, Y. Catalytic reaction mechanism of acetylcholinesterase determined by Born-Oppenheimer ab initio QM/MM molecular dynamics simulations. J. Phys. Chem. B 2010, 114, 8817–8825. [Google Scholar] [CrossRef] [PubMed]
- Sirin, G.S.; Zhou, Y.; Lior-Hoffmann, L.; Wang, S.; Zhang, Y. Aging mechanism of soman inhibited acetylcholinesterase. J. Phys. Chem. B 2012, 116, 12199–12207. [Google Scholar] [CrossRef] [PubMed]
- Sirin, G.S.; Zhang, Y. How is acetylcholinesterase phosphonylated by soman? An ab initio QM/MM molecular dynamics study. J. Phys. Chem. A 2014, 118, 9132–9139. [Google Scholar] [CrossRef] [PubMed]
- Enyedy, I.J.; Kovach, I.M.; Bencsura, A. Molecular dynamics study of active-site interactions with tetracoordinate transients in acetylcholinesterase and its mutants. Biochem. J. 2001, 353, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Cheng, X.; Radić, Z.; McCammon, J.A. Acetylcholinesterase: Mechanisms of covalent inhibition of wild-type and H447I mutant determined by computational analyses. J. Am. Chem. Soc. 2007, 129, 6562–6570. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.H.; Cheng, X.L.; Radić, Z.; McCammon, J.A. Acetylcholinesterase: Mechanisms of covalent inhibition of h447i mutant determined by computational analyses. Chem. Biol. Interact. 2008, 175, 196–199. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Cheng, S.; Sussman, J.L.; Silman, I.; Jiang, H. Computational Studies on Acetylcholinesterases. Molecules 2017, 22, 1324. https://doi.org/10.3390/molecules22081324
Xu Y, Cheng S, Sussman JL, Silman I, Jiang H. Computational Studies on Acetylcholinesterases. Molecules. 2017; 22(8):1324. https://doi.org/10.3390/molecules22081324
Chicago/Turabian StyleXu, Yechun, Shanmei Cheng, Joel L. Sussman, Israel Silman, and Hualiang Jiang. 2017. "Computational Studies on Acetylcholinesterases" Molecules 22, no. 8: 1324. https://doi.org/10.3390/molecules22081324




