Enantioselective Biosynthesis of l-Phenyllactic Acid by Whole Cells of Recombinant Escherichia coli
Abstract
:1. Introduction
2. Results
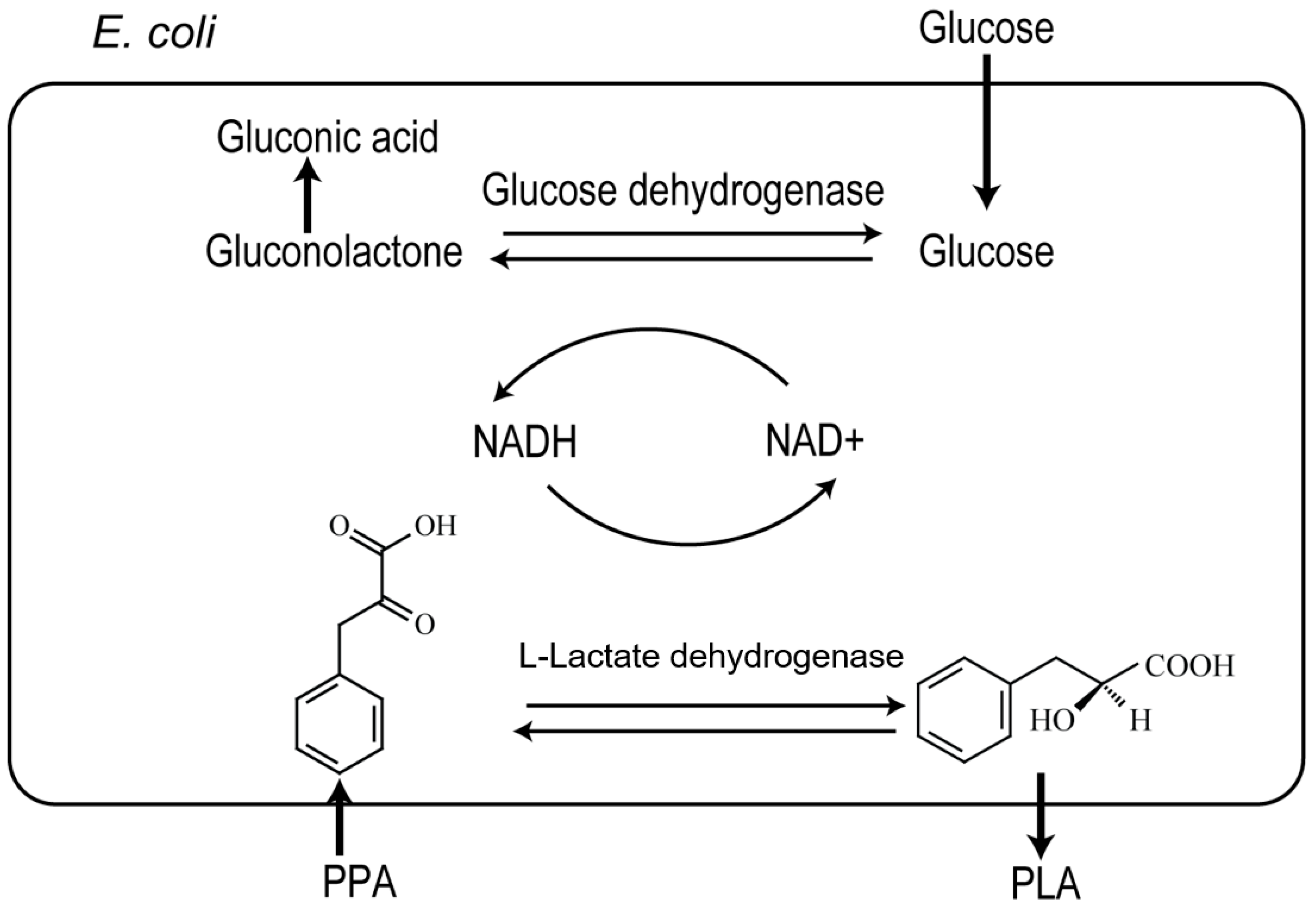
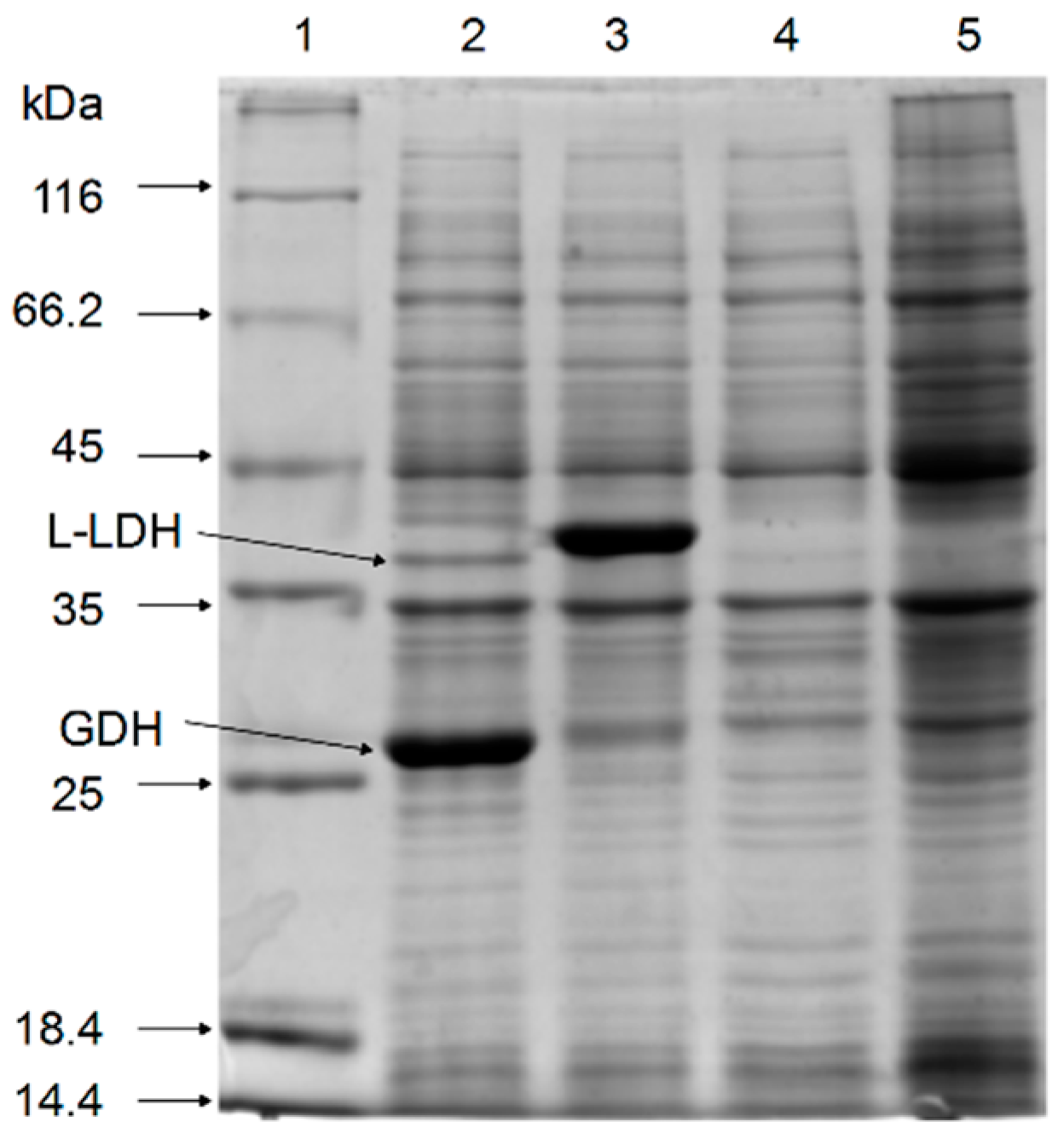
2.1. Cloning and Co-Expression of l-ldh and gdh in E. coli
2.2. Enzyme Assays of l-LDH and GDH
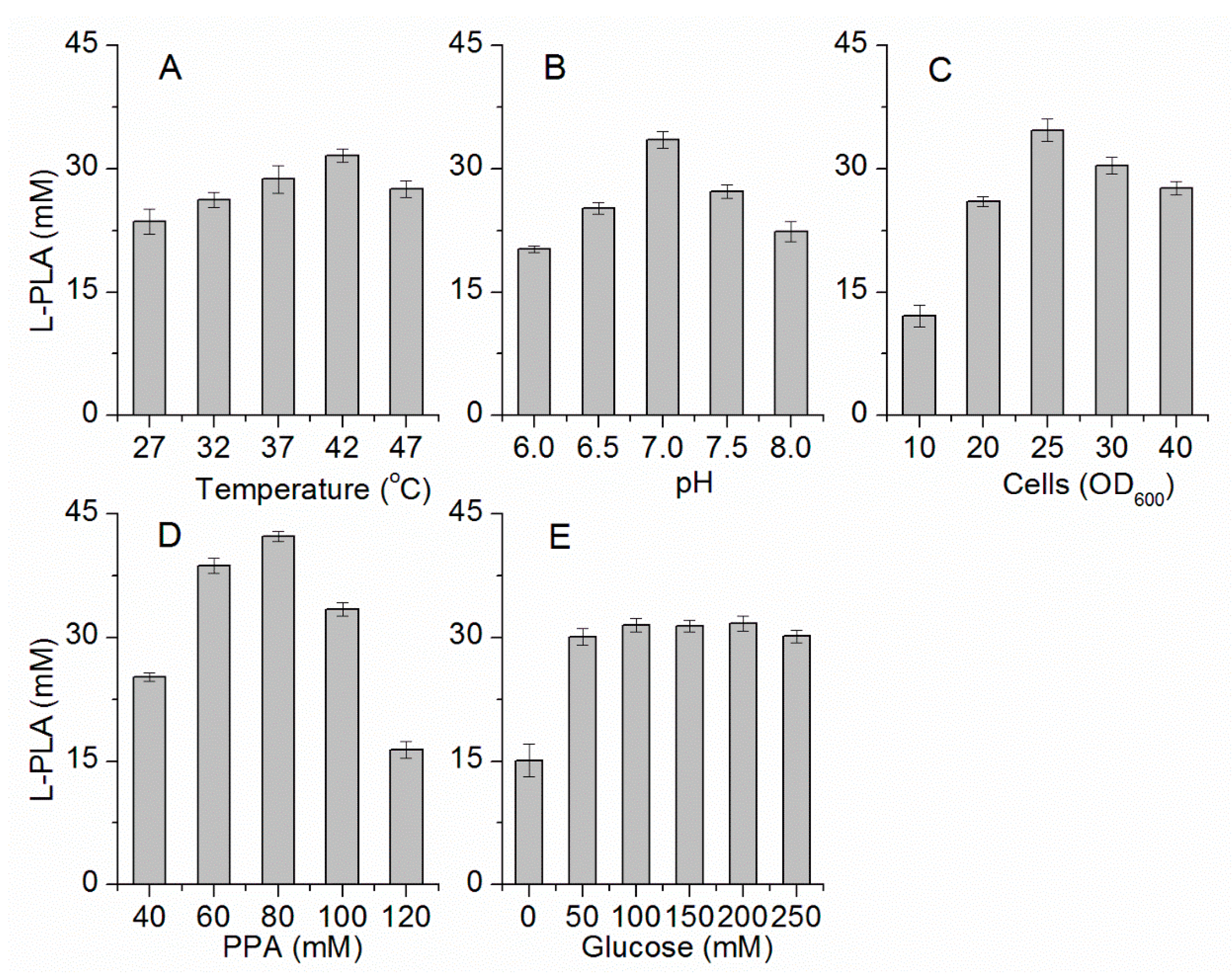
2.3. Optimization of Bioconversion Conditions
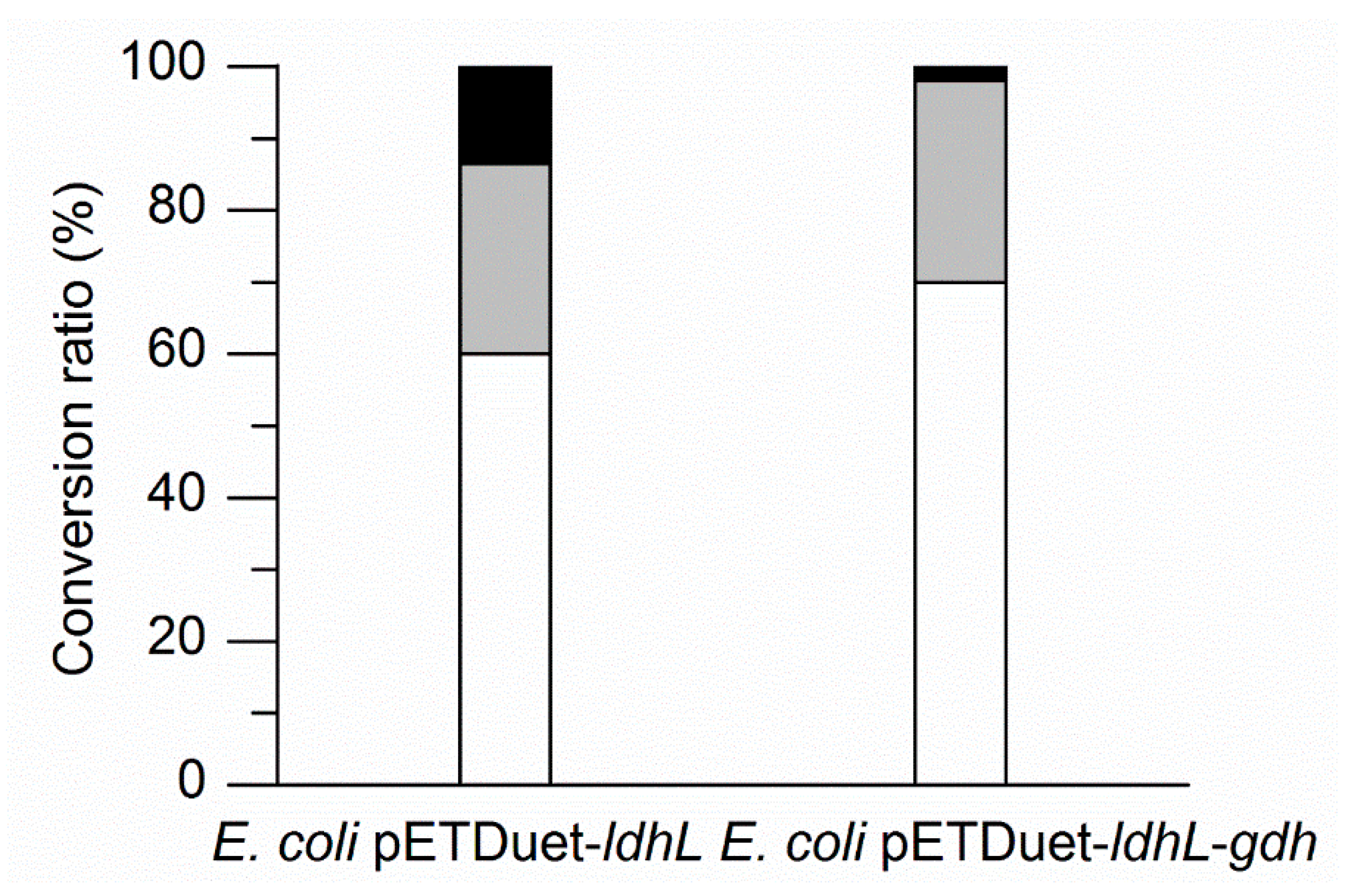
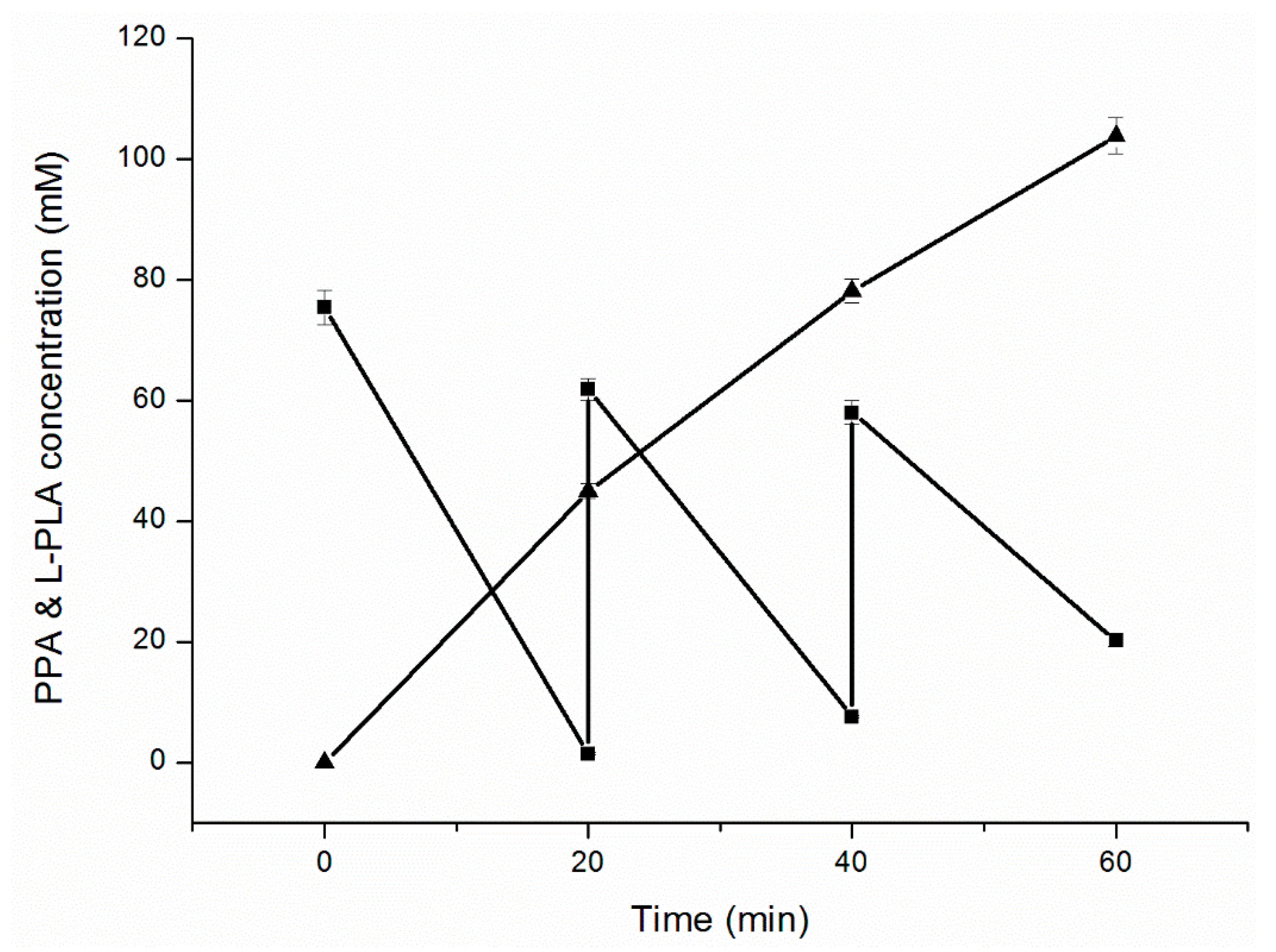
2.4. Enhancing l-PLA Production by a Cofactor Regeneration System
2.5. Fed-Batch Bioconversion of l-PLA
3. Discussion
4. Materials and Methods
4.1. Enzymes and Chemicals
4.2. Bacterial Strains, Plasmids, and Growth Conditions
4.3. Cloning and Expression of l-ldh and gdh in E. coli BL21(DE3)
4.4. Enzyme Assays
4.5. Biocatalyst Preparation and Optimization of Whole-Cell Biotransformation Conditions
4.6. Fed-Batch Bioconversion of l-PLA with Substrate Feeding by the Cofactor Regeneration System
4.7. Analytical Procedures
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tiwari, B.K.; Valdramidis, V.P.; O’Donnell, C.P.; Muthukumarappan, K.; Bourke, P.; Cullen, P. Application of natural antimicrobials for food preservation. J. Agric. Food Chem. 2009, 57, 5987–6000. [Google Scholar] [CrossRef] [PubMed]
- Crowley, S.; Mahony, J.; van Sinderen, D. Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives. Trends Food Sci. Technol. 2013, 33, 93–109. [Google Scholar] [CrossRef]
- Fujita, T.; Nguyen, H.D.; Ito, T.; Zhou, S.; Osada, L.; Tateyama, S.; Kaneko, T.; Takaya, N. Microbial monomers custom-synthesized to build true bio-derived aromatic polymers. Appl. Microbiol. Biotechnol. 2013, 97, 8887–8894. [Google Scholar] [CrossRef] [PubMed]
- Dieuleveux, V.; Lemarinier, S.; Gueguen, M. Antimicrobial spectrum and target site of d-3-phenyllactic acid. Int. J. Food Microbiol. 1998, 40, 177–183. [Google Scholar] [CrossRef]
- Schnurer, J.; Magnusson, J. Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci. Technol. 2005, 16, 70–78. [Google Scholar] [CrossRef]
- Lavermicocca, P.; Valerio, F.; Evidente, A.; Lazzaroni, S.; Corsetti, A.; Gobbetti, M. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl. Environ. Microbiol. 2000, 66, 4084–4090. [Google Scholar] [CrossRef] [PubMed]
- Ohhira, I.; Kuwaki, S.; Morita, H.; Suzuki, T.; Tomita, S.; Hisamatsu, S.; Sonoki, S.; Shinoda, S. Identification of 3-phenyllactic acid as a possible antibacterial substance produced by Enterococcus faecalis TH10. Biocontrol Sci. 2004, 9, 77–81. [Google Scholar] [CrossRef]
- Dieuleveux, V.; Guéguen, M. Antimicrobial effects of d-3-phenyllactic acid on Listeria monocytogenes in TSB-YE medium, milk, and cheese. J. Food Prot. 1998, 61, 1281–1285. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.K.; Kim, T.Y.; Park, S.J.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of polylactic acid and its copolymers. Biotechnol. Bioeng. 2010, 105, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Simmons, T.L.; Baker, G.L. Poly(phenyllactide): Synthesis, characterization, and hydrolytic degradation. Biomacromolecules 2001, 2, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Mu, W.; Zhang, T.; Jiang, B. Bioconversion of phenylpyruvate to phenyllactate: Gene cloning, expression, and enzymatic characterization of D- and L1-lactate dehydrogenases from Lactobacillus plantarum SK002. Appl. Biochem. Biotechnol. 2010, 162, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Kano, S.; Yuasa, Y.; Yokomatsu, T.; Shibuya, S. Highly stereocontrolled synthesis of the four individual stereoisomers of statine. J. Org. Chem. 1988, 53, 3865–3868. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Kobayashi, E.; Endo, T.; Nishiyama, M.; Horinouchi, S. Conversion of a cyanhydrin compound into S-(−)-3-phenyllactic acid by enantioselective hydrolytic activity of Pseudomonas sp. BC-18. Biosci. Biotechnol. Biochem. 1996, 60, 1279–1283. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.D.; Jin, X.; Kaneko, D.; Kaneko, T. Syntheses of high molecular weight poly (L-phenyllactic acid) s by a direct polycondensation in the presence of stable Lewis acids. Chem. Lett. 2011, 40, 584–585. [Google Scholar] [CrossRef]
- Ariens, E. Stereochemistry, a basis for sophisticated nonsense in pharmacokinetics and clinical pharmacology. Eur. J. Clin. Pharmacol. 1984, 26, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Bubl, E.C.; Butts, J.S. A method of synthesis of phenyllactic acid and substituted phenyllactic acids1. J. Am. Chem. Soc. 1951, 73, 4972. [Google Scholar] [CrossRef]
- Mu, W.M.; Liu, F.; Jia, J.; Chen, C.; Zhang, T.; Jiang, B. 3-Phenyllactic acid production by substrate feeding and pH-control in fed-batch fermentation of Lactobacillus sp. SK007. Bioresour. Technol. 2009, 100, 5226–5229. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, H.; Uematsu, K.; Ogino, C.; Teramura, H.; Niimi-Nakamura, S.; Tsuge, Y.; Hasunuma, T.; Oinuma, K.I.; Takaya, N.; Kondo, A. Simultaneous saccharification and fermentation of kraft pulp by recombinant Escherichia coli for phenyllactic acid production. Biochem. Eng. J. 2014, 88, 188–194. [Google Scholar] [CrossRef]
- Rodríguez-Pazo, N.; Vázquez-Araújo, L.; Pérez-Rodríguez, N.; Cortés-Diéguez, S.; Domínguez, J.M. Cell-free supernatants obtained from fermentation of cheese whey hydrolyzates and phenylpyruvic acid by Lactobacillus plantarum as a source of antimicrobial compounds, bacteriocins, and natural aromas. Appl. Biochem. Biotechnol. 2013, 171, 1042–1060. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Ma, C.; Gao, C.; Li, F.; Qin, J.; Zhang, H.; Wang, K.; Xu, P. Efficient conversion of phenylpyruvic acid to phenyllactic acid by using whole cells of Bacillus coagulans SDM. PLoS ONE 2011, 6, e19030. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.M.; Yu, S.; Zhu, L.; Zhang, T.; Jiang, B. Recent research on 3-phenyllactic acid, a broad-spectrum antimicrobial compound. Appl. Microbiol. Biotechnol. 2012, 95, 1155–1163. [Google Scholar] [CrossRef] [PubMed]
- Uppada, V.; Bhaduri, S.; Noronha, S.B. Cofactor regeneration—An important aspect of biocatalysis. Curr. Sci. India 2014, 106, 946–957. [Google Scholar]
- Li, X.; Jiang, B.; Pan, B.; Mu, W.; Zhang, T. Purification and partial characterization of Lactobacillus species SK007 lactate dehydrogenase (LDH) catalyzing phenylpyruvic acid (PPA) conversion into phenyllactic acid (PLA). J. Agric. Food Chem. 2008, 56, 2392–2399. [Google Scholar] [CrossRef] [PubMed]
- Prema, P.; Smila, D.; Palavesam, A.; Immanuel, G. Production and Characterization of an Antifungal Compound (3-Phenyllactic Acid) Produced by Lactobacillus plantarum Strain. Food Bioprocess Technol. 2010, 3, 379–386. [Google Scholar] [CrossRef]
- Strom, K.; Sjogren, J.; Broberg, A.; Schnurer, J. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(l-Phe-l-Pro) and cyclo(l-Phe-trans-4-OH-l-Pro) and 3-phenyllactic acid. Appl. Environ. Microbiol. 2002, 68, 4322–4327. [Google Scholar] [CrossRef] [PubMed]
- Valerio, F.; Lavermicocca, P.; Pascale, M.; Visconti, A. Production of phenyllactic acid by lactic acid bacteria: An approach to the selection of strains contributing to food quality and preservation. Fems Microbiol. Lett. 2004, 233, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hu, F.; Zhu, Y.; Wang, L.; Qi, B. Enhancement of phenyllactic acid biosynthesis by recognition site replacement of D-lactate dehydrogenase from Lactobacillus pentosus. Biotechnol. Lett. 2015, 37, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Xu, Y.; Wang, L.; Qi, B. Biosynthesis of (R)-2-hydroxy-3-phenylpropionic acid using whole recombinant Escherichia coli cells in an aqueous/n-octane biphasic system. J. Zhejiang Univ. Sci. B (Biomed. & Biotechnol.) 2017, in press. [Google Scholar] [CrossRef]
- Zhu, Y.; Jiang, Z.; Chen, J.; Xu, J.; Wang, L.; Qi, B. Fusion of D-Lactate Dehydrogenase and Formate Dehydrogenase for Increasing Production of (R)-3-Phenyllactic Acid in Recombinant Escherichia coli BL21 (DE3). J. Biobased Mater. Bioenergy 2017, 11, 372–378. [Google Scholar] [CrossRef]
- Prasuna, M.L.; Mujahid, M.; Sasikala, C.; Ramana, C.V. L-Phenylalanine catabolism and L-phenyllactic acid production by a phototrophic bacterium, Rubrivivax benzoatilyticus JA2. Microbiol. Res. 2012, 167, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.W.; Park, Y.; Penfound, T.; Fenger, T.; Spector, M. Regulation of NAD metabolism in Salmonella typhimurium: Molecular sequence analysis of the bifunctional nadR regulator and the nadA-pnuC operon. J. Bacteriol. 1990, 172, 4187–4196. [Google Scholar] [CrossRef] [PubMed]
- Berrı́, S.J.; Bennett, G.N.; San, K.Y. Metabolic engineering of Escherichia coli: Increase of NADH availability by overexpressing an NAD+-dependent formate dehydrogenase. Metab. Eng. 2002, 4, 217–229. [Google Scholar]
- Alissandratos, A.; Kim, H.K.; Easton, C.J. Formate production through carbon dioxide hydrogenation with recombinant whole cell biocatalysts. Bioresour. Technol. 2014, 164, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Hummel, W.; Kula, M.R. Dehydrogenases for the synthesis of chiral compounds. FEBS. J. 1989, 184, 85–97. [Google Scholar]
- Weckbecker, A.; Hummel, W. Glucose Dehydrogenase for the Regeneration of NADPH and NADH, in Microbial Enzymes and Biotransformations; Barredo, J.L., Ed.; Humana Press: New York, NY, USA, 2005; pp. 225–238. [Google Scholar]
- Goldberg, K.; Schroer, K.; Lütz, S.; Liese, A. Biocatalytic ketone reduction—A powerful tool for the production of chiral alcohols—Part I: Processes with isolated enzymes. Appl. Microbiol. Biotechnol. 2007, 76, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Zhao, M.; Zang, Y.; Zhou, Y.; Ouyang, J. Production of optically pure l-phenyllactic acid by using engineered Escherichia coli coexpressing l-lactate dehydrogenase and formate dehydrogenase. J. Biotechnol. 2015, 207, 45–71. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

| Strain | l-LDH Specific Activity (U mg−1) | GDH Specific Activity (U mg−1) |
|---|---|---|
| E. coli pETDuet-1 | ND | ND |
| E. coli pETDuet-ldhL | 28.11 ± 1.17 | ND |
| E. coli pETDuet-ldhL-gdh | 9.48 ± 0.91 | 5.37 ± 0.74 |
| Strain, Plasmid or Primer | Relevant Characteristics | Source |
|---|---|---|
| Lactobacillus plantarum | Wild type, source of l-ldh | CGMCC 1.2437 |
| Bacillus megaterium | Wild type, source of gdh | CCTCC M2013244 |
| E. coli DH5α | φ80 lacZΔM15Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 supE44λ-thi-1 | Novagen |
| E. coli BL21(DE3) | F− ompT hsdSB(rB−mB−)gal lonλdcm (DE3) | Novagen |
| E. coli pETDuet-1 | E. coli BL21(DE3) harboring pETDuet-1 | This study |
| E. coli pETDuet-ldhL-gdh | E. coli BL21(DE3) harboring pETDuet-ldhL-gdh | This study |
| E. coli pETDuet-ldhL | E. coli BL21(DE3) harboring pETDuet-ldhL | This study |
| pMD19-T (Simple) | Cloning vector, Amp r | TaKaRa |
| pMD-ldhL | l-ldh in pMD19-T | This study |
| pMD-gdh | gdh in pMD19-T | This study |
| pETDuet-1 | Expression vector, Amp r | Novagen |
| pETDuet-ldhL | l-ldh in pETDuet-1 | This study |
| pETDuet-ldhL-gdh | l-ldh and gdh in pETDuet-1 | This study |
| P1 (BamH I) | 5’-AAGGGATCCATTGTCAAGCATGCCAAATC-3’ | This study |
| P2 (Pst I) | 5’-CGCCTGCAGGGCCATTATTTATTTTCTAATTCAG-3’ | This study |
| P3 (Nde I) | 5’-GCGCCCATATGATGTATAAAGATTTAGAAGG-3’ | This study |
| P4 (Xho I) | 5’-CTAGCTCGAGCATTATCCGCGTCCTGCTT-3’ | This study |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Wang, Y.; Xu, J.; Chen, J.; Wang, L.; Qi, B. Enantioselective Biosynthesis of l-Phenyllactic Acid by Whole Cells of Recombinant Escherichia coli. Molecules 2017, 22, 1966. https://doi.org/10.3390/molecules22111966
Zhu Y, Wang Y, Xu J, Chen J, Wang L, Qi B. Enantioselective Biosynthesis of l-Phenyllactic Acid by Whole Cells of Recombinant Escherichia coli. Molecules. 2017; 22(11):1966. https://doi.org/10.3390/molecules22111966
Chicago/Turabian StyleZhu, Yibo, Ying Wang, Jiayuzi Xu, Jiahao Chen, Limei Wang, and Bin Qi. 2017. "Enantioselective Biosynthesis of l-Phenyllactic Acid by Whole Cells of Recombinant Escherichia coli" Molecules 22, no. 11: 1966. https://doi.org/10.3390/molecules22111966




