Production of Single-Chain Fv Antibodies Specific for GA-Pyridine, an Advanced Glycation End-Product (AGE), with Reduced Inter-Domain Motion
Abstract
:1. Introduction
2. Results
2.1. Preparation of Anti-GA-Pyridine scFvs by Phage Display
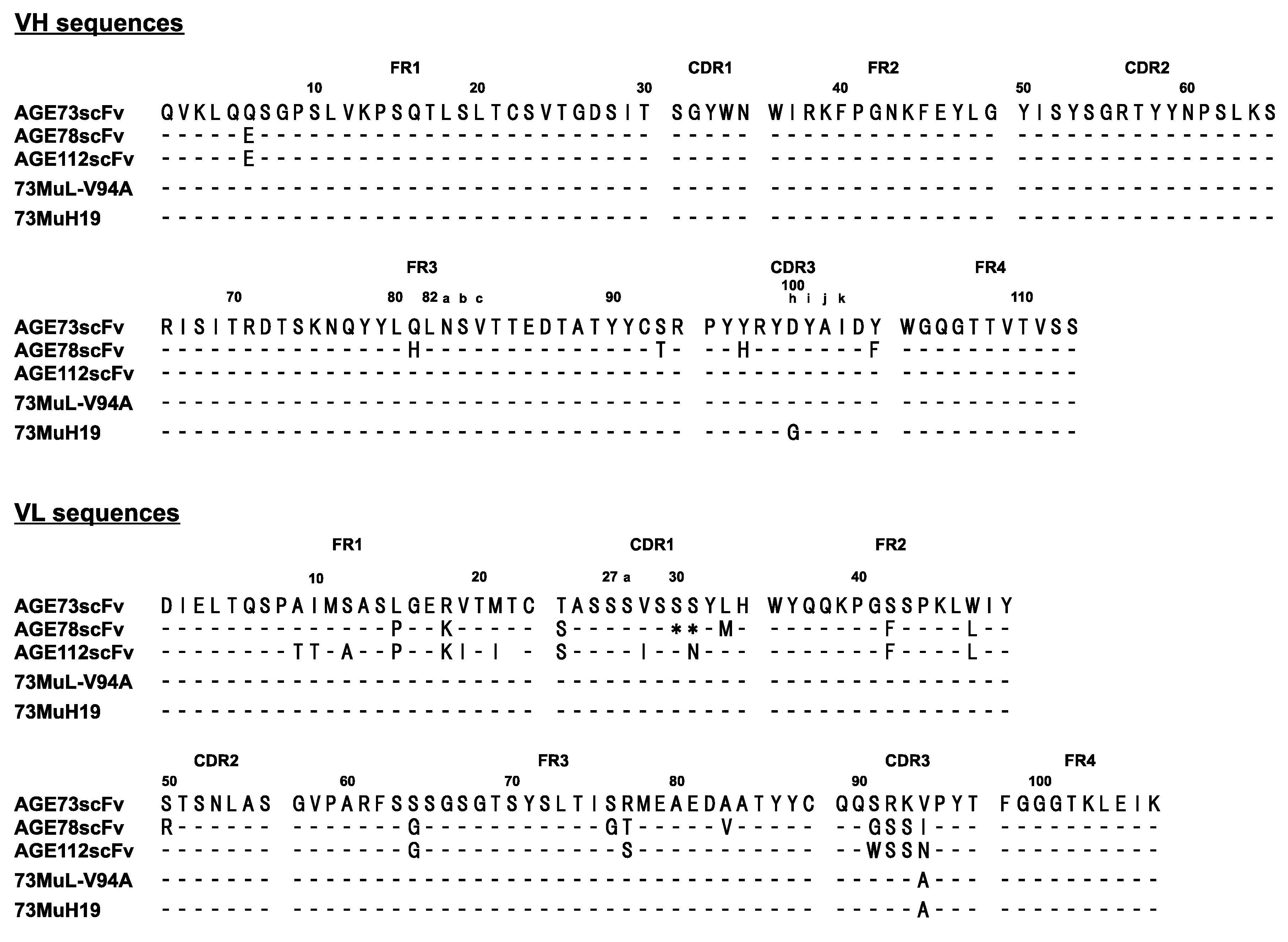
2.2. Selection of scFv with High Stability by Phage Display
2.3. Evaluation of Molecular Size by SAXS
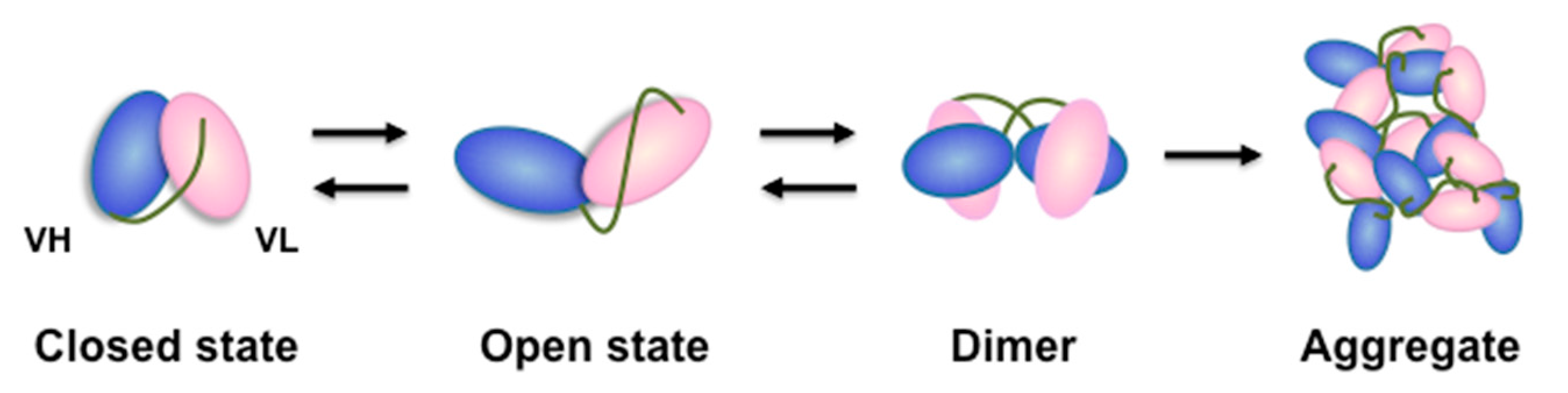
2.4. Direct Evaluation of Open–Close Dynamics by HS-AFM
2.5. NMR Measurements of 73MuH19
3. Discussion
4. Materials and Methods
4.1. Enzymes and Other Reagents
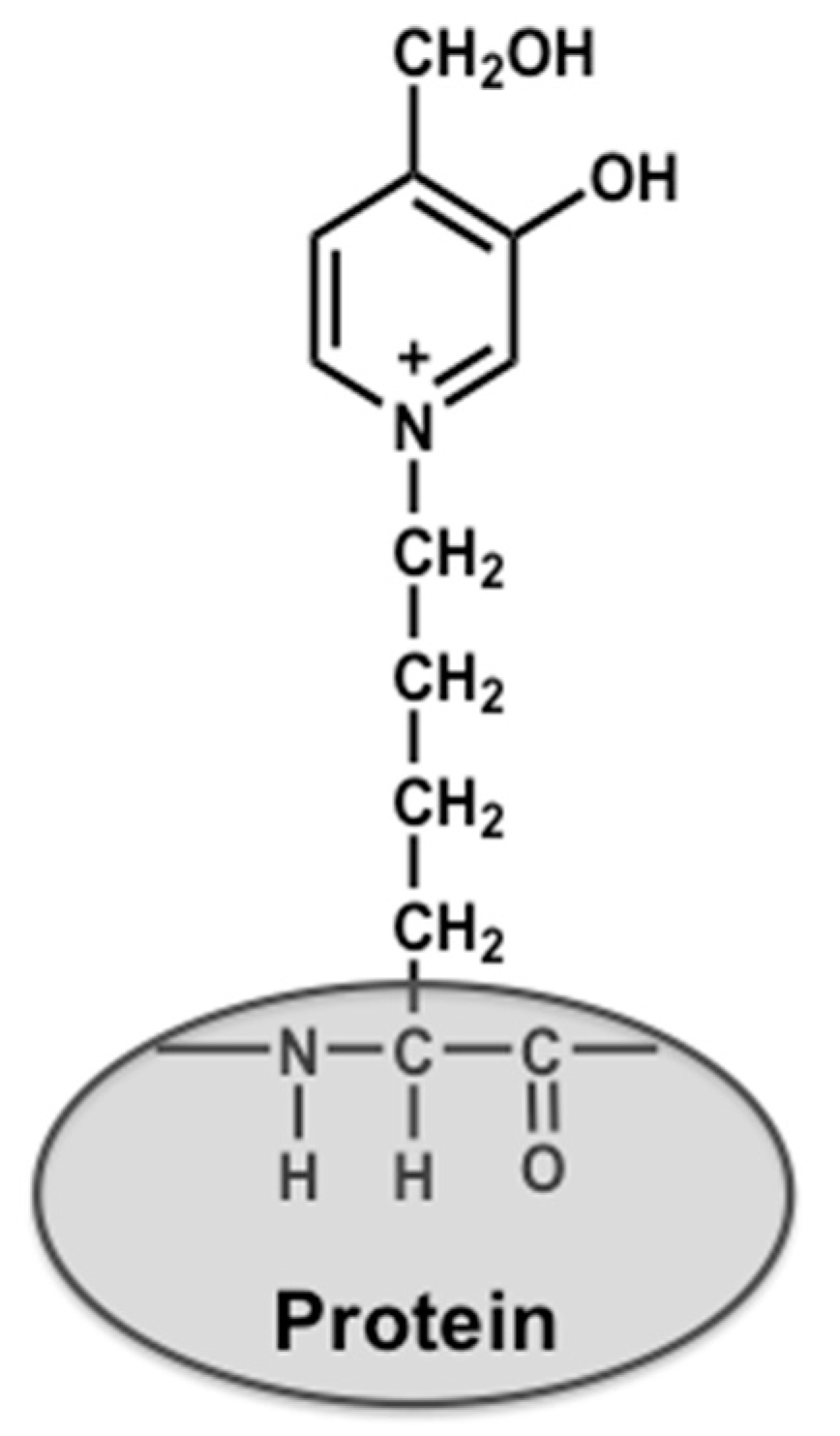
4.2. Preparation of Glycolaldehyde-Modified Bovine Serum Albumin (GA-BSA) and Nα-(Carbobenzyloxy)-GA-Pyridine (CBZ-GA-Pyridine)
4.3. Preparation of Biotinylated Peptide Containing GA-Pyridine
4.4. Selection of AGE73scFv by Phage Display
4.5. Phage Enzyme-Linked Immunosorbent Assay (ELISA)
4.6. Expression and Purification of scFv in E. coli
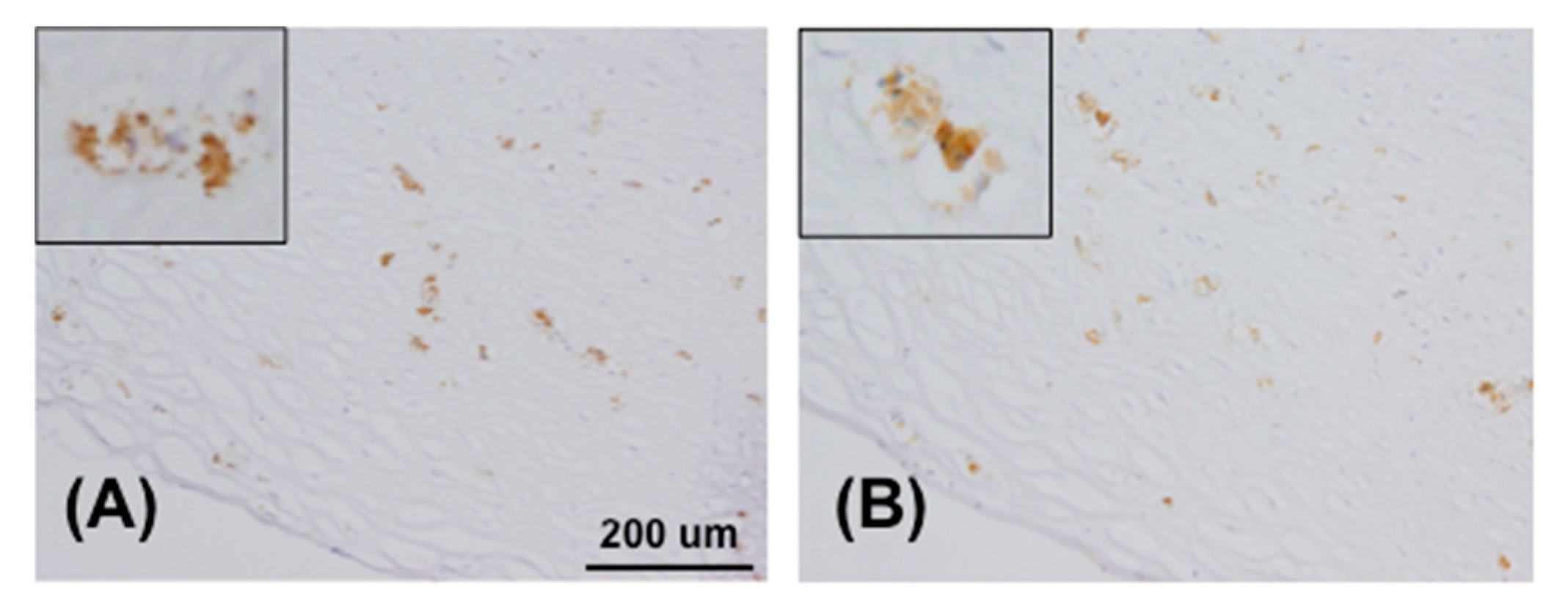
4.7. Immunohistochemistry
4.8. Surface Plasmon Resonance (SPR)
4.9. Construction of the Mutant scFv Library
4.10. Isothermal Titration Calorimetry (ITC)
4.11. Differential Scanning Calorimetry (DSC)
4.12. Small-Angle X-Ray Scattering (SAXS)
4.13. High-Speed Atomic Force Microscopy (HS-AFM)
4.14. NMR Spectroscopy
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Igawa, T.; Tsunoda, H.; Kuramochi, T.; Sampei, Z.; Ishii, S.; Hattori, K. Engineering the variable region of therapeutic IgG antibodies. MAbs 2011, 3, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Reichert, J.M. Marketed therapeutic antibodies compendium. MAbs 2012, 4, 413–415. [Google Scholar] [CrossRef] [PubMed]
- Ducancel, F.; Muller, B.H. Molecular engineering of antibodies for therapeutic and diagnostic purposes. MAbs 2012, 4, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Awazu, K.; Fujimaki, M.; Shimizu, K. Evaluation of anti-A/Udorn/307/1972 antibody specificity to influenza A/H3N2 viruses using an evanescent-field coupled waveguide-mode sensor. PLoS ONE 2013, 8, e81396. [Google Scholar] [CrossRef] [PubMed]
- Plückthun, A.; Glockshuber, R.; Pfitzinger, I.; Skerra, A.; Stadlmüller, J. Engineering of antibodies with a known three-dimensional structure. Cold Spring Harb. Symp. Quant. Biol. 1987, 52, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Huston, J.S.; Levinson, D.; Mudgett-Hunter, M.; Tai, M.S.; Novotný, J.; Margolies, M.N.; Ridge, R.J.; Bruccoleri, R.E.; Haber, E.; Crea, R.; et al. Protein engineering of antibody binding sites: Recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. USA 1988, 85, 5879–5883. [Google Scholar] [CrossRef] [PubMed]
- Plückthun, A.; Skerra, A. Expression of functional antibody Fv and Fab fragments in Escherichia coli. Methods Enzymol. 1989, 178, 497–515. [Google Scholar] [PubMed]
- Plückthun, A. Antibody engineering: Advances from the use of Escherichia coli expression systems. Biotechnology 1991, 9, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Desplancq, D.; King, D.J.; Lawson, A.D.; Mountain, A. Multimerization behaviour of single chain Fv variants for the tumour-binding antibody B72.3. Protein Eng. 1994, 7, 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.C.; Boehm, M.K.; Chester, K.A.; Begent, R.H.; Perkins, S.J. Reversible dimer formation and stability of the anti-tumour single-chain Fv antibody MFE-23 by neutron scattering, analytical ultracentrifugation, and NMR and FT-IR spectroscopy. J. Mol. Biol. 2002, 320, 107–127. [Google Scholar] [CrossRef]
- Wilkinson, I.C.; Hall, C.J.; Veverka, V.; Shi, J.Y.; Muskett, F.W.; Stephens, P.E.; Taylor, R.J.; Henry, A.J.; Carr, M.D. High resolution NMR-based model for the structure of a scFv-IL-1β complex: Potential for NMR as a key tool in therapeutic antibody design and development. J. Biol. Chem. 2009, 284, 31928–31935. [Google Scholar] [CrossRef] [PubMed]
- Kortt, A.A.; Malby, R.L.; Caldwell, J.B.; Gruen, L.C.; Ivancic, N.; Lawrence, M.C.; Howlett, G.J.; Webster, R.G.; Hudson, P.J.; Colman, P.M. Recombinant anti-sialidase single-chain variable fragment antibody. Characterization, formation of dimer and higher-molecular-mass multimers and the solution of the crystal structure of the single-chain variable fragment/sialidase complex. Eur. J. Biochem. 1994, 221, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Arndt, K.M.; Müller, K.M.; Plückthun, A. Factors influencing the dimer to monomer transition of an antibody single-chain Fv fragment. Biochemistry 1998, 37, 12918–12926. [Google Scholar] [CrossRef] [PubMed]
- Glockshuber, R.; Malia, M.; Pfitzinger, I.; Plückthun, A. A comparison of strategies to stabilize immunoglobulin Fv-fragments. Biochemistry 1990, 29, 1362–1367. [Google Scholar] [CrossRef] [PubMed]
- Young, N.M.; MacKenzie, C.R.; Narang, S.A.; Oomen, R.P.; Baenziger, J.E. Thermal stabilization of a single-chain Fv antibody fragment by introduction of a disulphide bond. FEBS Lett. 1995, 377, 135–139. [Google Scholar] [CrossRef]
- Dooley, H.; Grant, S.D.; Harris, W.J.; Porter, A.J. Stabilization of antibody fragments in adverse environments. Biotechnol. Appl. Biochem. 1998, 28, 77–83. [Google Scholar] [PubMed]
- Wörn, A.; Plückthun, A. Different equilibrium stability behavior of scFv fragments: Identification, classification, and improvement by protein engineering. Biochemistry 1999, 38, 8739–8750. [Google Scholar] [CrossRef] [PubMed]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Honegger, A.; Plückthun, A. Selection for improved protein stability by phage display. J. Mol. Biol. 1999, 294, 163–180. [Google Scholar] [CrossRef] [PubMed]
- Brockmann, E.C.; Cooper, M.; Strömsten, N.; Vehniäinen, M.; Saviranta, P. Selecting for antibody scFv fragments with improved stability using phage display with denaturation under reducing conditions. J. Immunol. Methods 2005, 296, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Nagai, R.; Hayashi, C.M.; Xia, L.; Takeya, M.; Horiuchi, S. Identification in human atherosclerotic lesions of GA-pyridine, a novel structure derived from glycolaldehyde-modified proteins. J. Biol. Chem. 2002, 277, 48905–48912. [Google Scholar] [CrossRef] [PubMed]
- Hirose, J.; Yamabe, S.; Takada, K.; Okamoto, N.; Nagai, R.; Mizuta, H. Immunohistochemical distribution of advanced glycation end products (AGEs) in human osteoarthritic cartilage. Acta Histochem. 2011, 113, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Svergun, D.I.; Barberato, C.; Koch, M.H. CRYSOL—A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995, 28, 768–773. [Google Scholar] [CrossRef]
- Suzuki, N.; Ichikawa, N.; Kasai, S.; Yamada, M.; Nishi, N.; Morioka, H.; Yamashita, H.; Kitagawa, Y.; Utani, A.; Hoffman, M.P.; et al. Syndecan binding site in the laminin alpha1 chain G domain. Biochemistry 2003, 42, 12625–12633. [Google Scholar] [CrossRef] [PubMed]
- Miyata, K.; Takagi, S.; Sato, S.; Morioka, H.; Shiba, K.; Minamisawa, T.; Takami, M.; Fujita, N. Suppression of Aggrus/podoplanin-induced platelet aggregation and pulmonary metastasis by a single-chain antibody variable region fragment. Cancer Med. 2014, 3, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Morioka, H.; Tobisawa, K.; Torizawa, T.; Kato, K.; Shimada, I.; Nikaido, O.; Stewart, J.D.; Ohtsuka, E. Probing the interaction between a high-affinity single-chain Fv and a pyrimidine (6-4) pyrimidone photodimer by site-directed mutagenesis. Biochemistry 1999, 38, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, N.; Suwa, Y.; Uchida, M.; Kobashigawa, Y.; Yokoyama, H.; Morioka, H. Role of the mobility of antigen binding site in high affinity antibody elucidated by surface plasmon resonance. J. Biochem. 2017, 161, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Kobashigawa, Y.; Saio, T.; Ushio, M.; Sekiguchi, M.; Yokochi, M.; Ogura, K.; Inagaki, F. Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination. J. Biomol. NMR 2012, 53, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Krishnarjuna, B.; Jaipuria, G.; Thakur, A.; D'Silva, P.; Atreya, H.S. Amino acid selective unlabeling for sequence specific resonance assignments in proteins. J. Biomol. NMR 2010, 49, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Waugh, D.S. Genetic tools for selective labeling of proteins with alpha-l5N-amino acids. J. Biomol. NMR 1996, 8, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Tsuboki, J.; Fujiwara, Y.; Horlad, H.; Shiraishi, D.; Nohara, T.; Tayama, S.; Motohara, T.; Saito, Y.; Ikeda, T.; Takaishi, K.; et al. Onionin A inhibits ovarian cancer progression by suppressing cancer cell proliferation and the protumour function of macrophages. Sci. Rep. 2016, 6, 29588. [Google Scholar] [CrossRef] [PubMed]
- Kobashigawa, Y.; Fukuda, N.; Nakahara, Y.; Morioka, H. Surface plasmon resonance. In Advanced Methods in Structural Biology; Senda, T., Maenaka, K., Eds.; Springer: Tokyo, Japan, 2016; pp. 227–237. [Google Scholar]
- Leung, D.W.; Chen, E.Y.; Goeddel, D.V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1989, 1, 11–15. [Google Scholar]
- Cadwell, R.C.; Joyce, G.F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992, 2, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Beckman, R.A.; Mildvan, A.S.; Loeb, L.A. On the fidelity of DNA replication: Manganese mutagenesis in vitro. Biochemistry 1985, 24, 5810–5815. [Google Scholar] [CrossRef] [PubMed]
- Petoukhov, M.V.; Franke, D.; Shkumatov, A.V.; Tria, G.; Kikhney, A.G.; Gajda, M.; Gorba, C.; Mertens, H.D.T.; Konarev, P.V.; Svergun, D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 2012, 45, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Guinier, A.; Fournet, G. Small-Angle Scattering of X-ray; John Wiley & Sons: New York, NY, USA, 1955. [Google Scholar]
- Ando, T.; Kodera, N.; Takai, E.; Maruyama, D.; Saito, K.; Toda, A. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. USA 2001, 98, 12468–12472. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Uchihashi, T.; Kodera, N.; Yamamoto, D.; Miyagi, A.; Taniguchi, M.; Yamashita, H. High-speed AFM and nano-visualization of biomolecular processes. Pflüg. Arch. Eur. J. Physiol. 2008, 456, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Wendel, M.; Lorenz, H.; Kotthaus, J.P. Sharpened electron beam deposited tips for high resolution atomic force microscope lithography and imaging. Appl. Phys. Lett. 1995, 67, 3723–3734. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
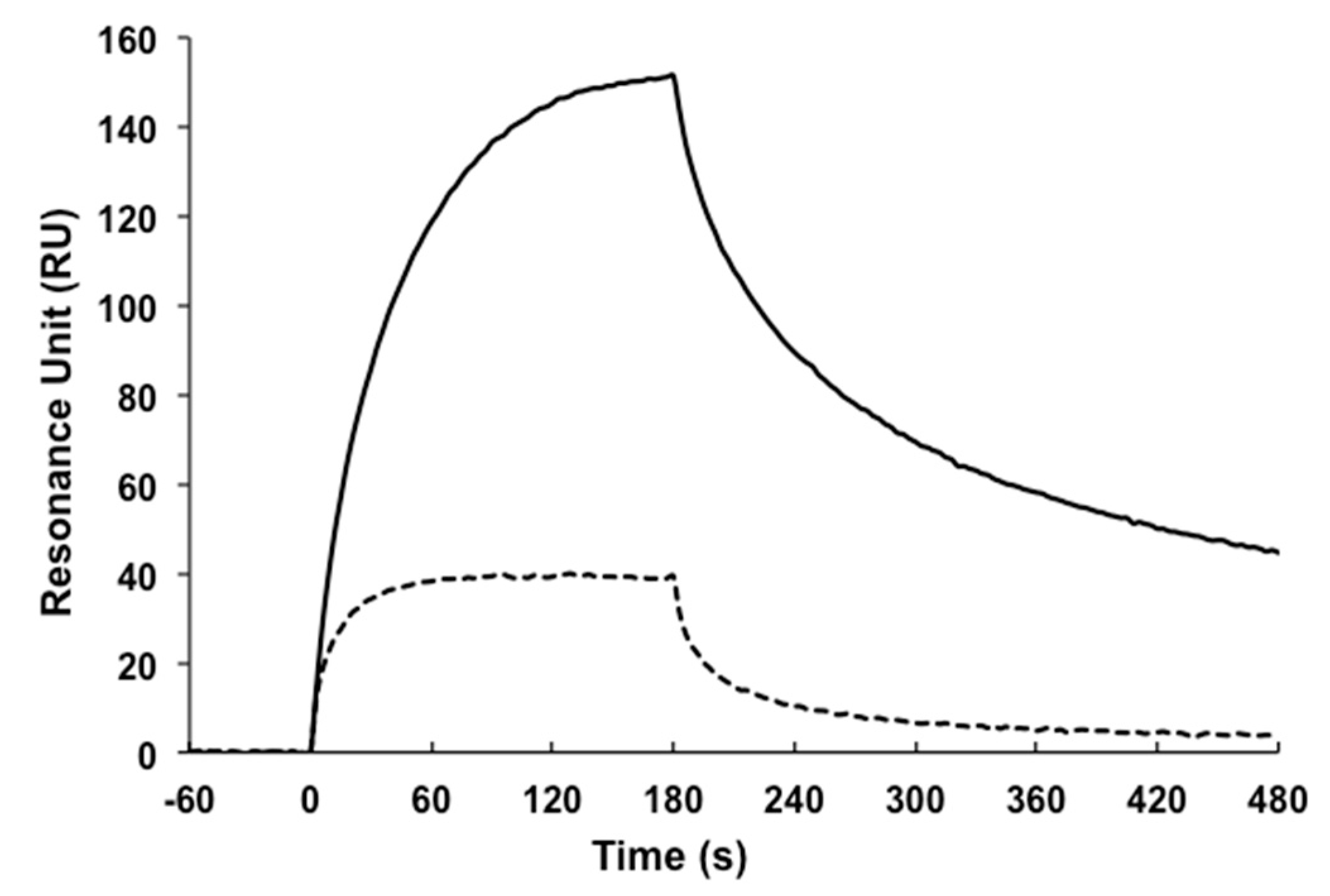
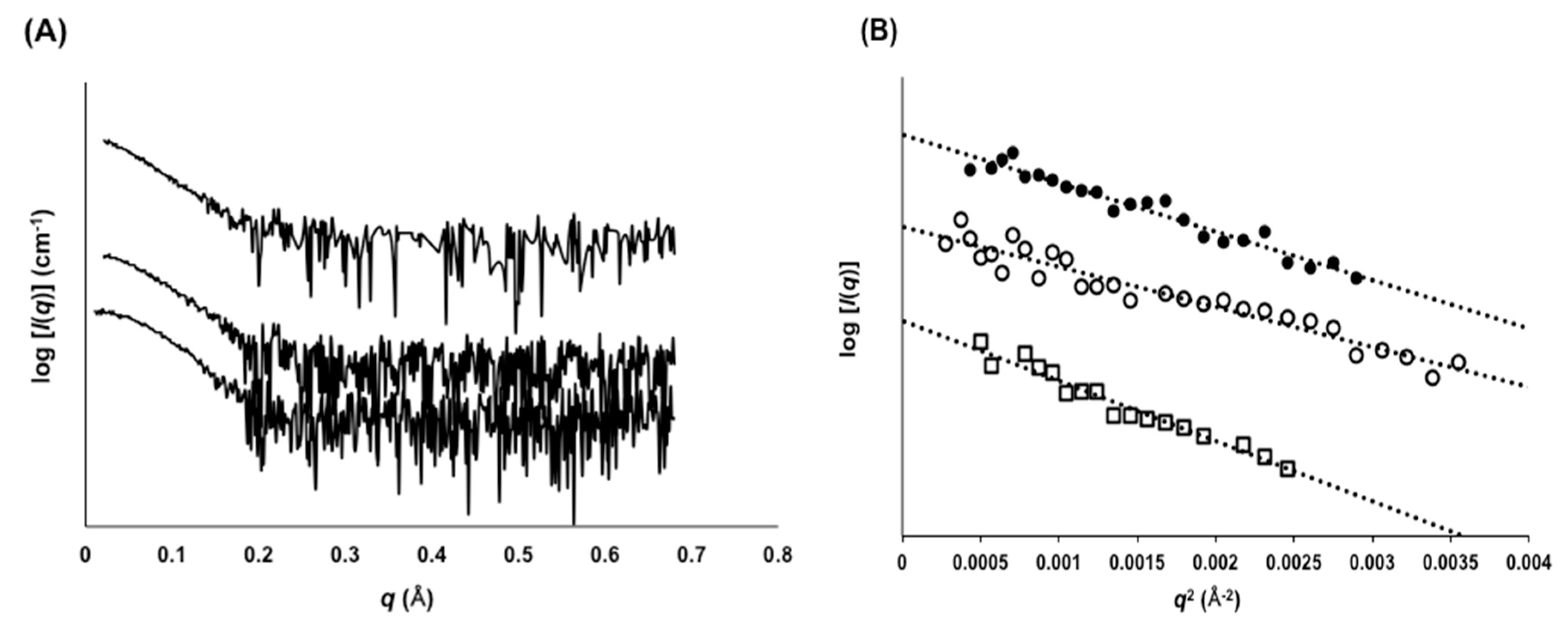
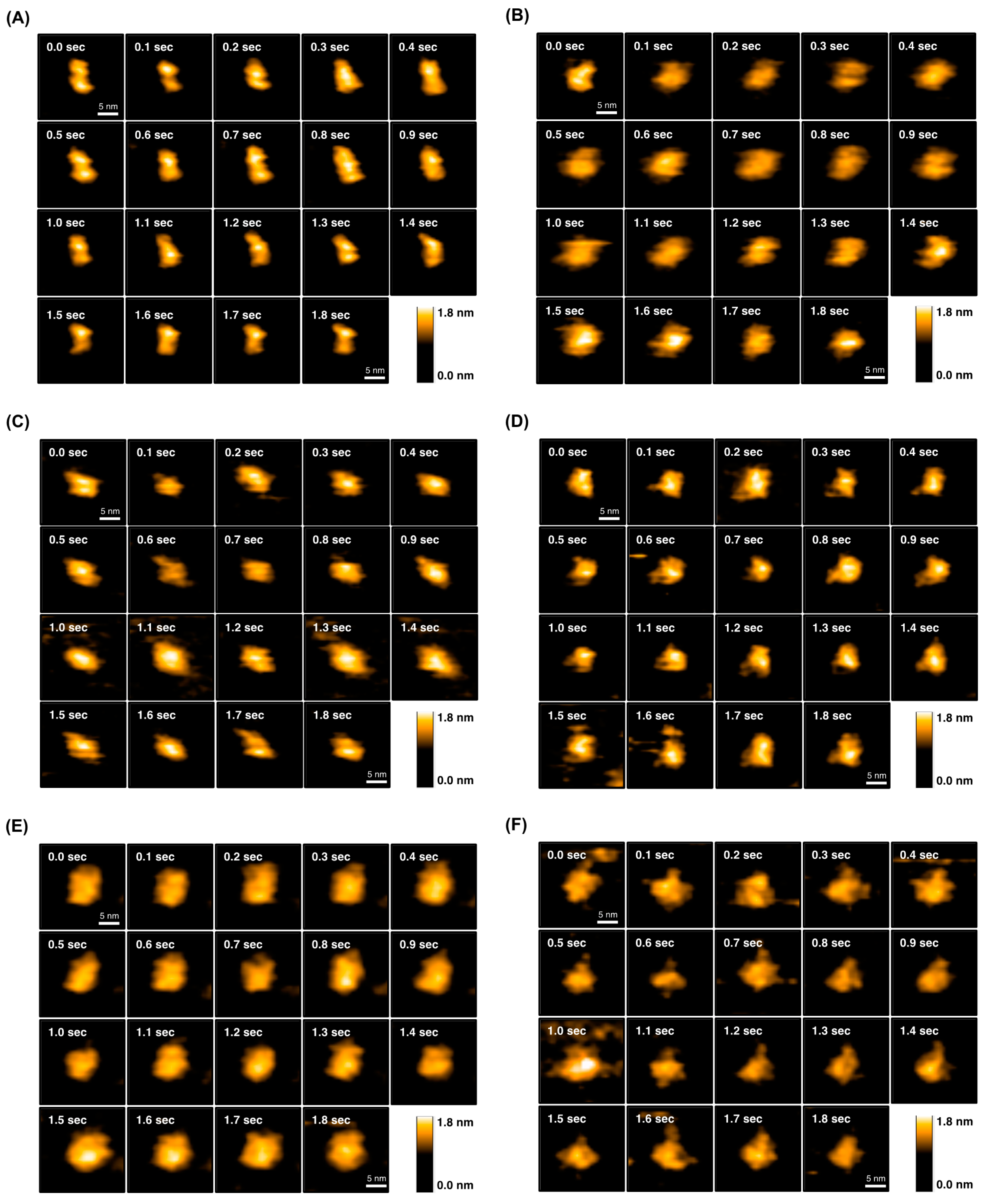

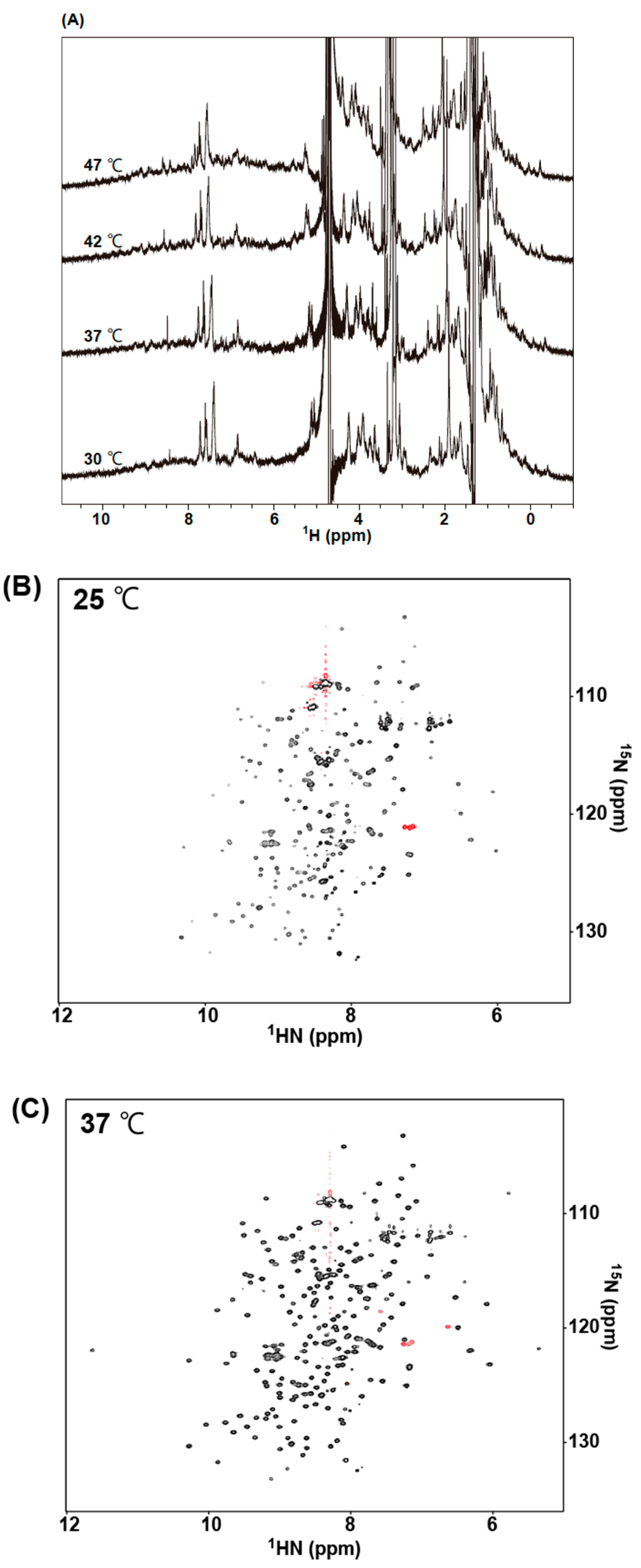
| scFv Clones | 1 KD × 10−8 (M) | 2 Tm (°C) | 3 Rg (Å) | |
|---|---|---|---|---|
| 25 °C | 37 °C | |||
| AGE73scFv | 13.4 ± 0.5 | 68.4 ± 13.2 | 62.8 ± 0.02 | 25.1 ± 0.5 |
| 73MuL-V94A | 5.3 ± 1.3 | 14.9 ± 2.7 | 62.7 ± 0.02 | 22.5 ± 0.6 |
| 73MuH19 | 4.7 ± 1.1 | 23.0 ± 5.3 | 65.0 ± 0.02 | 20.5 ± 0.8 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukuda, N.; Noi, K.; Weng, L.; Kobashigawa, Y.; Miyazaki, H.; Wakeyama, Y.; Takaki, M.; Nakahara, Y.; Tatsuno, Y.; Uchida-Kamekura, M.; et al. Production of Single-Chain Fv Antibodies Specific for GA-Pyridine, an Advanced Glycation End-Product (AGE), with Reduced Inter-Domain Motion. Molecules 2017, 22, 1695. https://doi.org/10.3390/molecules22101695
Fukuda N, Noi K, Weng L, Kobashigawa Y, Miyazaki H, Wakeyama Y, Takaki M, Nakahara Y, Tatsuno Y, Uchida-Kamekura M, et al. Production of Single-Chain Fv Antibodies Specific for GA-Pyridine, an Advanced Glycation End-Product (AGE), with Reduced Inter-Domain Motion. Molecules. 2017; 22(10):1695. https://doi.org/10.3390/molecules22101695
Chicago/Turabian StyleFukuda, Natsuki, Kentaro Noi, Lidong Weng, Yoshihiro Kobashigawa, Hiromi Miyazaki, Yukari Wakeyama, Michiyo Takaki, Yusuke Nakahara, Yuka Tatsuno, Makiyo Uchida-Kamekura, and et al. 2017. "Production of Single-Chain Fv Antibodies Specific for GA-Pyridine, an Advanced Glycation End-Product (AGE), with Reduced Inter-Domain Motion" Molecules 22, no. 10: 1695. https://doi.org/10.3390/molecules22101695
APA StyleFukuda, N., Noi, K., Weng, L., Kobashigawa, Y., Miyazaki, H., Wakeyama, Y., Takaki, M., Nakahara, Y., Tatsuno, Y., Uchida-Kamekura, M., Suwa, Y., Sato, T., Ichikawa-Tomikawa, N., Nomizu, M., Fujiwara, Y., Ohsaka, F., Saitoh, T., Maenaka, K., Kumeta, H., ... Morioka, H. (2017). Production of Single-Chain Fv Antibodies Specific for GA-Pyridine, an Advanced Glycation End-Product (AGE), with Reduced Inter-Domain Motion. Molecules, 22(10), 1695. https://doi.org/10.3390/molecules22101695





