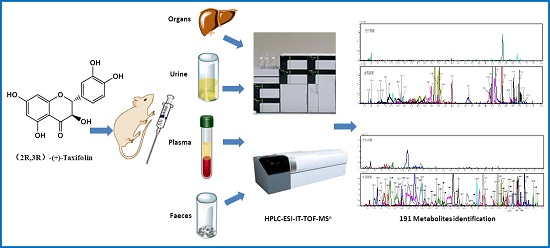
Detection of 191 Taxifolin Metabolites and Their Distribution in Rats Using HPLC-ESI-IT-TOF-MSn
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Taxifolin in Rats and Study on the Fragmentation Behaviours of Taxifolin and Reference Compounds
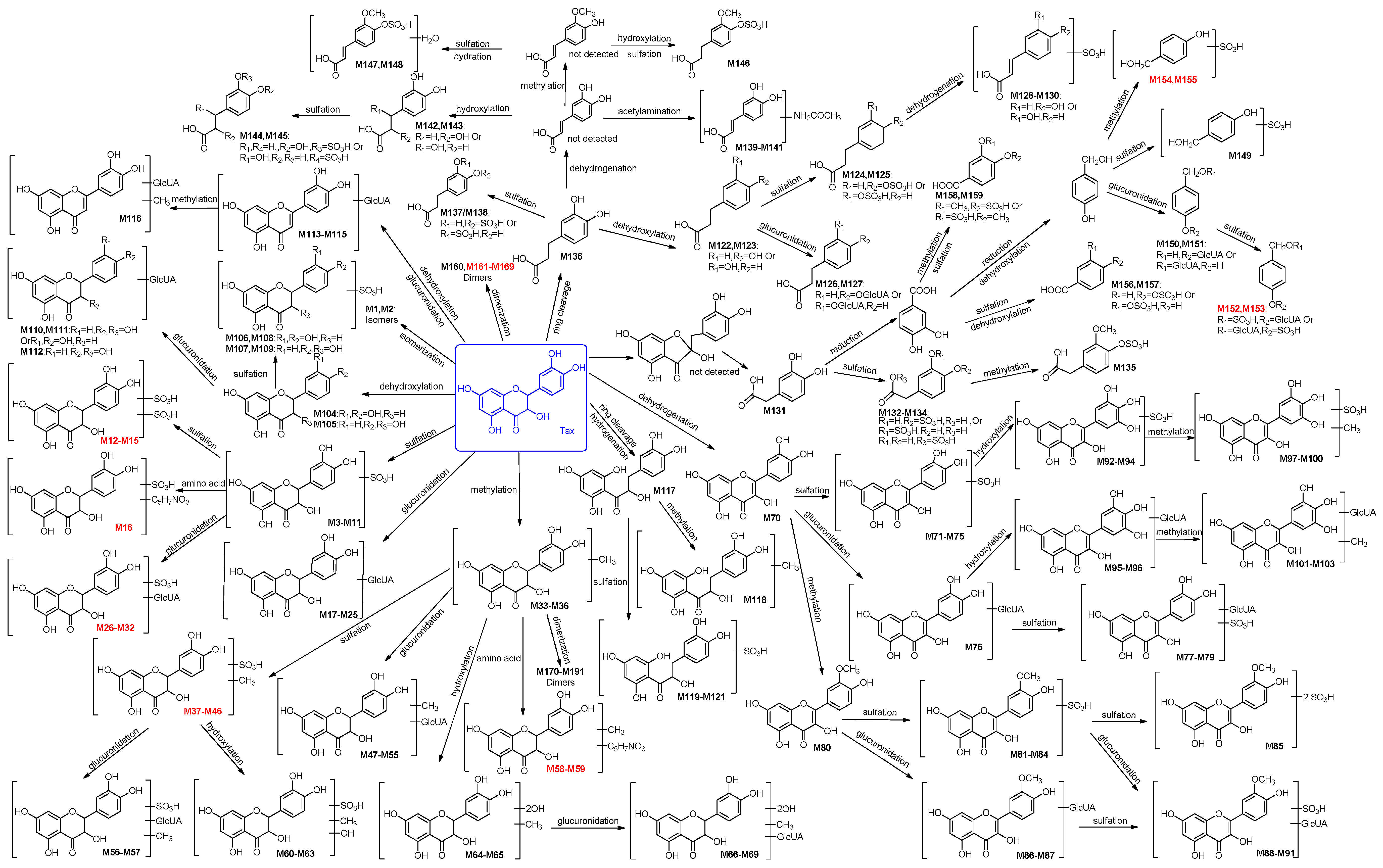
2.2. Identification of 191 Metabolites of Taxifolin in Rats
2.2.1. Identification of 32 Metabolites (M1–M32) Having the Aglycone of Taxifolin or Its Isomers
2.2.2. Identification of 37 Metabolites (M33–M69) Having the Aglycone of Methyl Taxifolin
2.2.3. Identification of 34 Metabolites (M70–M103) Having the Aglycone of Quercetin
2.2.4. Identification of 9 Metabolites (M104–M112) Having the Aglycone of Dehydroxylated Taxifolin
2.2.5. Identification of Four Metabolites (M113–M116) Formed through Dehydration and Glucuronidation
2.2.6. Identification of Five Metabolites (M117–M121) Having the Aglycone of Hydrogenated Taxifolin
2.2.7. Identification of 38 Metabolites (M122–M159) Having the Aglycone of Phenolic Acid Derivatives
2.2.8. Identification of 32 Metabolites (M160–M191) Formed through Dimerization
2.3. Distribution of the Metabolites of Taxifolin in Rats
2.4. Bioactivities of the Metabolites of Taxifolin
2.5. Prediction of Taxifolin Metabolite Targets
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Animals and Drug Administration
3.3. Urine and Faeces Samples Collection and Preparation
3.4. Blood Sample Collection and Preparation
3.5. Organ Sample Collection and Preparation
3.6. Instruments and Conditions for HPLC-ESI-IT-TOF-MSn
3.7. Prediction of Taxifolin Metabolite Targets
3.8. Determination of the Level of Identification for All Metabolites
- Level 1:
- The metabolites are identified by comparison with reference compounds.
- Level 2:
- The metabolites are identified by comparison with reference literature or can be found in the Scifinder database.
- Level 3:
- New metabolites/compounds that could not be found in the SciFinder database.
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vrhovsek, U.; Masuero, D.; Gasperotti, M.; Franceschi, P.; Caputi, L.; Viola, R.; Mattivi, F. A versatile targeted metabolomics method for the rapid quantification of multiple classes of phenolics in fruits and beverages. J. Agric. Food. Chem. 2012, 60, 8831–8840. [Google Scholar] [CrossRef] [PubMed]
- Schauss, A.G.; Tselyico, S.S.; Kuznetsova, V.A.; Yegorova, I. Toxicological and genotoxicity assessment of a dihydroquercetin-rich dahurian larch tree (Larix gmelinii Rupr) extract (Lavitol). Int. J. Toxicol. 2015, 34, 162–181. [Google Scholar] [CrossRef] [PubMed]
- Slimestad, R.; Fossen, T.; Vagen, I.M. Onions: a source of unique dietary flavonoids. J. Agric. Food. Chem. 2007, 55, 10067–10080. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.N.; Carrier, D.J.; Clausen, E.C. Batch solvent extraction of flavanolignans from milk thistle (Silybum marianum L. Gaertner). Phytochem. Anal. 2005, 16, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Trebaticka, J.; Kopasova, S.; Hradecna, Z.; Cinovsky, K.; Skodacek, I.; Suba, J.; Muchova, J.; Zitnanova, I.; Waczulikova, I.; Rohdewald, P.; et al. Treatment of ADHD with French maritime pine bark extract, Pycnogenol. Eur. Child Adoles. Psy. 2006, 15, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Kiehlmann, E.; Li, E.P.M. Isomerization of dihydroquercetin. J. Nat. Prod. 1995, 58, 450–455. [Google Scholar] [CrossRef]
- Vega-Villa, K.R.; Remsberg, C.M.; Ohgami, Y.; Yanez, J.A.; Takemoto, J.K.; Andrews, P.K.; Davies, N.M. Stereospecific high-performance liquid chromatography of taxifolin, applications in pharmacokinetics, and determination in tu fu ling (Rhizoma Smilacis glabrae) and apple (Malus x domestica). Biomed. Chromatogr. 2009, 23, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Weidmann, A.E. Dihydroquercetin: More than just an impurity? Eur. J. Pharmacol. 2012, 684, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Topal, F.; Nar, M.; Gocer, H.; Kalin, P.; Kocyigit, U.M.; Gulcin, I.; Alwasel, S.H. Antioxidant activity of taxifolin: an activity-structure relationship. J. Enzyme Inhib. Med. Chem. 2016, 31, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Willfoer, S.M.; Ahotupa, M.O.; Hemming, J.E.; Reunanen, M.H.T.; Eklund, P.C.; Sjoeholm, R.E.; Eckerman, C.S.E.; Pohjamo, S.P.; Holmbom, B.R. Antioxidant activity of knotwood extractives and phenolic compounds of selected tree species. J. Agric. Food. Chem. 2003, 51, 7600–7606. [Google Scholar] [CrossRef] [PubMed]
- Oi, N.; Chen, H.; Kim, M.O.; Lubet, R.A.; Bode, A.M.; Dong, Z. Taxifolin suppresses UV-induced skin carcinogenesis by targeting EGFR and PI3K. Cancer Prev. Res. 2012, 5, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Delporte, C.; Backhouse, N.; Erazo, S.; Negrete, R.; Vidal, P.; Silva, X.; Lopez-Perez, J.L.; Feliciano, A.S.; Munoz, O. Analgesic-antiinflammatory properties of Proustia pyrifolia. J. Ethnopharmacol. 2005, 99, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Gocer, H.; Topal, F.; Topal, M.; Kucuk, M.; Teke, D.; Gulcin, I.; Alwasel, S.H.; Supuran, C.T. Acetylcholinesterase and carbonic anhydrase isoenzymes I and II inhibition profiles of taxifolin. J. Enzyme Inhib. Med. Chem. 2015, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Fedosova, N.F.; Alisievich, S.V.; Lyadov, K.V.; Romanova, E.P.; Rud′Ko, I.A.; Kubatiev, A.A. Mechanisms underlying diquertin-mediated regulation of neutrophil function in patients with non-insulin-dependent diabetes mellitus. Bull. Exp. Biol. Med. 2004, 137, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Chen, R.C.; Yang, Z.H.; Sun, G.B.; Wang, M.; Ma, X.J.; Yang, L.J.; Sun, X.B. Taxifolin prevents diabetic cardiomyopathy in vivo and in vitro by inhibition of oxidative stress and cell apoptosis. Food Chem. Toxicol. 2014, 63, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Ezzikouri, S.; Nishimura, T.; Kohara, M.; Benjelloun, S.; Kino, Y.; Inoue, K.; Matsumori, A.; Tsukiyama-Kohara, K. Inhibitory effects of pycnogenol on hepatitis C virus replication. Antiviral Res. 2015, 113, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Fongang, Y.S.F.; Bankeu, J.J.K.; Ali, M.S.; Awantu, A.F.; Zeeshan, A.; Assob, C.N.; Mehreen, L.; Lenta, B.N.; Ngouela, S.A.; Tsamo, E. Flavonoids and other bioactive constituents from Ficus thonningii Blume (Moraceae). Phytochem. Lett. 2015, 11, 139–145. [Google Scholar] [CrossRef]
- Zhao, M.; Chen, J.; Zhu, P.; Fujino, M.; Takahara, T.; Toyama, S.; Tomita, A.; Zhao, L.; Yang, Z.; Hei, M.; et al. Dihydroquercetin (DHQ) ameliorated concanavalin A-induced mouse experimental fulminant hepatitis and enhanced HO-1 expression through MAPK/Nrf2 antioxidant pathway in RAW cells. Int. Immunopharmacol. 2015, 28, 938–944. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, X.; Cui, Y.; Zhou, H.; Xu, D.; Shan, T.; Zhang, F.; Guo, Y.; Chen, Y.; Wu, D. Taxifolin protects against cardiac hypertrophy and fibrosis during biomechanical stress of pressure overload. Toxicol. Appl. Pharmacol. 2015, 287, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Dok-Go, H.; Lee, K.H.; Kim, H.J.; Lee, E.H.; Lee, J.; Song, Y.S.; Lee, Y.H.; Jin, C.; Lee, Y.S.; Cho, J. Neuroprotective effects of antioxidative flavonoids, quercetin, (+)-dihydroquercetin and quercetin 3-methyl ether, isolated from Opuntia ficus-indica var. saboten. Brain Res. 2003, 965, 130–136. [Google Scholar] [CrossRef]
- Ahn, J.Y.; Choi, S.E.; Jeung, M.S.; Park, K.H.; Moon, N.J.; Joo, S.S.; Lee, C.S.; Choi, Y.W.; Li, K.; Lee, M.K.; et al. Effect of taxifolin glycoside on atopic dermatitis-like skin lesions in NC/Nga mice. Phytother. Res. 2010, 24, 1071–1077. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Gao, C.; Luo, M.; Wang, W.; Zhao, C.; Zu, Y.; Efferth, T.; Fu, Y. Dihydroquercetin (DHQ) induced HO-1 and NQO1 expression against oxidative stress through the Nrf2-dependent antioxidant pathway. J. Agric. Food Chem. 2013, 61, 2755–2761. [Google Scholar] [CrossRef] [PubMed]
- Booth, A.N.; DeEds, F. The toxicity and metabolism of dihydroquercetin. J. Am. Pharm. Assoc. Am. Pharm. Assoc. 1958, 47, 183–184. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.P.; Abd-el-Mohsen, M.M.; Rice-Evans, C. Cellular uptake and metabolism of flavonoids and their metabolites: Implications for their bioactivity. Arch. Biochem. Biophys. 2004, 423, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Jokura, H.; Hashizume, K.; Ominami, H.; Shibuya, Y.; Suzuki, A.; Hase, T.; Shimotoyodome, A. Hesperidin metabolite hesperetin-7-O-glucuronide, but not hesperetin-3′-O-glucuronide, exerts hypotensive, vasodilatory, and anti-inflammatory activities. Food Funct. 2013, 4, 1346–1351. [Google Scholar] [CrossRef] [PubMed]
- Lotito, S.B.; Zhang, W.-J.; Yang, C.-S.; Crozier, A.; Frei, B. Metabolic conversion of dietary flavonoids alters their anti-inflammatory and antioxidant properties. Free Radic. Biol. Med. 2011, 51, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Wiczkowski, W.; Szawara-Nowak, D.; Topolska, J.; Olejarz, K.; Zieliński, H.; Pisku A, M.K. Metabolites of dietary quercetin: Profile, isolation, identification, and antioxidant capacity. J. Funct. Foods 2014, 11, 121–129. [Google Scholar] [CrossRef]
- Tang, H.; Tang, L.; Xu, R.; Yang, J. Phase II metabolites of several flavonoids and their pharmacological activities. Zhongguo Xin Yao Za Zhi 2012, 21, 144–150. [Google Scholar]
- Kawabata, K.; Mukai, R.; Ishisaka, A. Quercetin and related polyphenols: new insights and implications for their bioactivity and bioavailability. Food Funct. 2015, 6, 1399–1417. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D. Studies on the Transport of Taxifolin and Astilbin in Vitro and Pharmacokinetics in Rats. Ph.D. Thesis, Zhejiang University, Zhejiang, China, 2009. [Google Scholar]
- Duweler, K.G.; Rohdewald, P. Urinary metabolites of French maritime pine bark extract in humans. Pharmazie 2000, 55, 364–368. [Google Scholar] [PubMed]
- Liang, J.; Xu, F.; Zhang, Y.Z.; Zang, X.Y.; Wang, D.; Shang, M.Y.; Wang, X.; Chui, D.H.; Cai, S.Q. The profiling and identification of the metabolites of (+)-catechin and study on their distribution in rats by HPLC-DAD-ESI-IT-TOF-MSn technique. Biomed. Chromatogr. 2014, 28, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.R.; Xu, F.; Li, D.P.; Lu, F.L.; Liu, G.X.; Wang, L.; Shang, M.Y.; Huang, Y.L.; Cai, S.Q. Metabolites of Siamenoside I and their distributions in rats. Molecules 2016, 21, 176. [Google Scholar] [CrossRef] [PubMed]
- Blaut, M.; Schoefer, L.; Braune, A. Transformation of flavonoids by intestinal microorganisms. Int. J. Vitam. Nutr. Res. 2003, 73, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Vacek, J.; Papouskova, B.; Kosina, P.; Vrba, J.; Kren, V.; Ulrichova, J. Biotransformation of flavonols and taxifolin in hepatocyte in vitro systems as determined by liquid chromatography with various stationary phases and electrospray ionization-quadrupole time-of-flight mass spectrometry. J. Chromatogr. B 2012, 899, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Meng, M.X.; Gao, L.B.; Liu, T.; Xu, Q.; Zeng, S. Permeation of astilbin and taxifolin in Caco-2 cell and their effects on the P-gp. Int. J. Pharm. 2009, 378, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Xia, H.J.; Xing, F.; Deng, G.F.; Shen, Q.; Zeng, S. A highly sensitive and robust UPLC-MS with electrospray ionization method for quantitation of taxifolin in rat plasma. J. Chromatogr. B 2009, 877, 1778–1786. [Google Scholar] [CrossRef] [PubMed]
- Pozharitskaya, O.N.; Karlina, M.V.; Shikov, A.N.; Kosman, V.M.; Makarova, M.N.; Makarov, V.G. Determination and pharmacokinetic study of taxifolin in rabbit plasma by high-performance liquid chromatography. Phytomedicine 2009, 16, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Xu, F.; Zhang, Y.Z.; Huang, S.; Zang, X.Y.; Zhao, X.; Zhang, L.; Shang, M.Y.; Yang, D.H.; Wang, X.; et al. The profiling and identification of the absorbed constituents and metabolites of paeoniae radix rubra decoction in rat plasma and urine by the HPLC-DAD-ESI-IT-TOF-MSn technique: a novel strategy for the systematic screening and identification of absorbed constituents and metabolites from traditional Chinese medicines. J. Pharm. Biomed. Anal. 2013, 83, 108–121. [Google Scholar] [PubMed]
- Ma, L.M.; Xu, F.; Li, F.C.; Wang, J.Z.; Shang, M.Y.; Liu, G.X.; Cai, S.Q. The profiling and identification of the metabolites of 8-prenylkaempferol and a study on their distribution in rats by high-performance liquid chromatography with diode array detection combined with electrospray ionization ion trap time-of-flight multistage mass spectrometry. Biomed. Chromatogr. 2015, 30, 175–190. [Google Scholar] [PubMed]
- Van der Woude, H.; Boersma, M.G.; Vervoort, J.; Rietjens, I.M. Identification of 14 quercetin phase II mono- and mixed conjugates and their formation by rat and human phase II in vitro model systems. Chem. Res. Toxicol. 2004, 17, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- Marmet, C.; Actis-Goretta, L.; Renouf, M.; Giuffrida, F. Quantification of phenolic acids and their methylates, glucuronides, sulfates and lactones metabolites in human plasma by LC-MS/MS after oral ingestion of soluble coffee. J. Pharm. Biomed. Anal. 2014, 88, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Uutela, P.; Reinila, R.; Harju, K.; Piepponen, P.; Ketola, R.A.; Kostiainen, R. Analysis of intact glucuronides and sulfates of serotonin, dopamine, and their phase I metabolites in rat brain microdialyzates by liquid chromatography-tandem mass spectrometry. Anal. Chem. 2009, 81, 8417–8425. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.L.; Dai, Y.; Yao, Z.H.; Qin, Z.F.; Wang, X.L.; Qin, L.; Yao, X.S. Metabolites profile of Xian-Ling-Gu-Bao capsule, a traditional Chinese medicine prescription, in rats by ultra performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry analysis. J. Pharm. Biomed. Anal. 2014, 96, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Redeuil, K.; Smarrito-Menozzi, C.; Guy, P.; Rezzi, S.; Dionisi, F.; Williamson, G.; Nagy, K.; Renouf, M. Identification of novel circulating coffee metabolites in human plasma by liquid chromatography-mass spectrometry. J. Chromatogr. A 2011, 1218, 4678–4688. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.; Kotnik, P.; Trontelj, J.; Knez, Ž.; Mašič, L.P. Bioactivation of bisphenol A and its analogs (BPF, BPAF, BPZ and DMBPA) in human liver microsomes. Toxicol. In Vitro 2013, 27, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Xu, F.; Dong, J.; Liang, J.; Hashi, Y.; Shang, M.Y.; Yang, D.H.; Wang, X.; Cai, S.Q. Profiling and identification of the metabolites of calycosin in rat hepatic 9000 x g supernatant incubation system and the metabolites of calycosin-7-O-β-d-glucoside in rat urine by HPLC-DAD-ESI-IT-TOF-MSn technique. J. Pharm. Biomed. Anal. 2012, 70, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Tang, M.; Song, H.; Li, R.; Wang, C.; Ye, H.; Qiu, N.; Zhang, Y.; Chen, L.; Wei, Y. Characterization of metabolic profile of honokiol in rat feces using liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry and 13C stable isotope labeling. J. Chromatogr. B 2014, 953–954, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Contreras, J.; Sippl, W. Homo and heterodimer ligands: the twin drug approach. In The Practice of Medicinal Chemistry, 3rd ed.; Wermuth, C.G., Ed.; Elsevier Ltd.: London, UK, 2008; pp. 380–383. [Google Scholar]
- Purchartova, K.; Valentova, K.; Pelantova, H.; Marhol, P.; Cvacka, J.; Havlicek, L.; Krenkova, A.; Vavrikova, E.; Biedermann, D.; Chambers, C.S.; et al. Prokaryotic and Eukaryotic Aryl Sulfotransferases: Sulfation of Quercetin and Its Derivatives. Chemcatchem 2015, 7, 3152–3162. [Google Scholar] [CrossRef]
- Yoshizumi, M.; Tsuchiya, K.; Kirima, K.; Kyaw, M.; Suzaki, Y.; Tamaki, T. Quercetin inhibits Shc- and phosphatidylinositol 3-kinase-mediated c-Jun N-terminal kinase activation by angiotensin II in cultured rat aortic smooth muscle cells. Mol. Pharmacol. 2001, 60, 656–665. [Google Scholar] [PubMed]
- Kim, J.E.; Lee, D.E.; Lee, K.W.; Son, J.E.; Seo, S.K.; Li, J.; Jung, S.K.; Heo, Y.S.; Mottamal, M.; Bode, A.M. Isorhamnetin suppresses skin cancer through direct inhibition of MEK1 and PI3-K. Cancer Prev. Res. 2011, 4, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Nicolau, M.; Dovichi, S.S.; Cuttle, G. Pro-inflammatory effect of quercetin by dual blockade of angiotensin converting-enzyme and neutral endopeptidase in vivo. Nutr. Neurosci. 2003, 6, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.L.; Rupasinghe, S.G.; Felicia, S.; Schuler, M.A.; Elvira, G.D.M. Citrus flavonoids luteolin, apigenin, and quercetin inhibit glycogen synthase kinase-3β enzymatic activity by lowering the interaction energy within the binding cavity. J. Med. Food 2011, 14, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Boussoualim, N.; Abderrahmane, B. Kinetic study of different flavonoids as inhibitors of beta-lactamase enzyme. Afr. J. Biochem. Res. 2011, 321–327. [Google Scholar]
- Song, K.S.; Choi, S.H.; Hur, J.M.; Park, H.J.; Yang, E.J.; Mook-Jung, I.; Yi, J.H.; Jun, M. Inhibitory effects of flavonoids isolated from leaves of Petasites japonicus on β-secretase (BACE1). Food Sci. Biotechnol. 2008, 17, 1165–1170. [Google Scholar]
- Wan, Y.; Tang, M.H.; Chen, X.C.; Chen, L.J.; Wei, Y.Q.; Wang, Y.S. Inhibitory effect of liposomal quercetin on acute hepatitis and hepatic fibrosis induced by concanavalin A. Braz. J. Med. Biol. Res. 2014, 47, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Sawai, Y. NMR analytical approach to clarify the molecular mechanisms of the antioxidative and radical-scavenging activities of antioxidants in tea: Reaction of polyphenols with stable free radicals. Yasai Chagyo Kenkyusho Kenkyu Hokoku 2007, 6, 23–58. [Google Scholar]
- Duenas, M.; Surco-Laos, F.; Gonzalez-Manzano, S.; Gonzalez-Paramas, A.M.; Santos-Buelga, C. Antioxidant properties of major metabolites of quercetin. Eur. Food Res. Technol. 2011, 232, 103–111. [Google Scholar] [CrossRef]
- Yagi, A.; Uemura, T.; Okamura, N.; Haraguchi, H.; Imoto, T.; Hashimoto, K. Antioxidative sulfated flavonoids in leaves of Polygonum hydropiper. Phytochemistry 1994, 35, 885–887. [Google Scholar] [CrossRef]
- Seo, K.; Yang, J.H.; Kim, S.C.; Ku, S.K.; Ki, S.H.; Shin, S.M. The antioxidant effects of isorhamnetin contribute to inhibit COX-2 expression in response to inflammation: a potential role of HO-1. Inflammation 2014, 37, 712–722. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.K.; Song, K.S.; Kim, C.J.; Kim, H.M.; Oh, G.T.; Yoo, I.D. Tumor cell growth inhibition and antioxidant activity of flavonoids from the stem bark of Cudrania tricuspidata. Han′guk Nonghwa Hakhoechi 1994, 37, 105–109. [Google Scholar]
- Boesch-Saadatmandi, C.; Loboda, A.; Wagner, A.E.; Stachurska, A.; Jozkowicz, A.; Dulak, J.; Doring, F.; Wolffram, S.; Rimbach, G. Effect of quercetin and its metabolites isorhamnetin and quercetin-3-glucuronide on inflammatory gene expression: role of miR-155. J. Nutr. Biochem. 2011, 22, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Ksouri, R.; Ksouri, W.M.; Jallali, I.; Debez, A.; Magne, C.; Hiroko, I.; Abdelly, C. Medicinal halophytes: potent source of health promoting biomolecules with medical, nutraceutical and food applications. Crit. Rev. Biotechnol. 2012, 32, 289–326. [Google Scholar] [CrossRef] [PubMed]
- Armah, F.A.; Annan, K.; Mensah, A.Y.; Amponsah, I.K.; Tocher, D.A.; Habtemariam, S. Erythroivorensin: A novel anti-inflammatory diterpene from the root-bark of Erythrophleum ivorense (A Chev.). Fitoterapia 2015, 105, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Liu, C.; Chen, R. Phenolic constituents from stem bark of Morus wittiorum and their anti-inflammation and cytotoxicity. Zhongguo Zhong Yao Za Zhi 2010, 35, 2700–2703. [Google Scholar] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.Q.; Wang, X.; Shang, M.Y.; Xu, F.; Liu, G.X. “Efficacy theory” may help to explain characteristic advantages of traditional Chinese medicines. Zhongguo Zhong Yao Za Zhi 2015, 40, 3435–3443. [Google Scholar] [PubMed]
- Li, C.; Xu, F.; Xie, D.M.; Jing, Y.; Shang, M.Y.; Liu, G.X.; Wang, X.; Cai, S.Q. Identification of absorbed constituents in the rabbit plasma and cerebrospinal fluid after intranasal administration of Asari Radix et Rhizoma by HS-SPME-GC-MS and HPLC-APCI-IT-TOF-MSn. Molecules 2014, 19, 4857–4879. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are not available from the authors.
| No. | tR (min) | Formula | Ion | Meas. m/z | Pred. m/z | Diff (ppm) | DBE | Urine | Plasma | Faeces | Identification Level | Identification |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TAX | 41.023 | C15H12O7 | [M − H]− | 303.0521 | 303.0510 | 3.63 | 10 | ▲ | ▲ | ▲ | Level 1 | Taxifolin (parent compound) |
| Metabolites having the aglycone of taxifolin or its isomers (M1–M32); two bioactive metabolites | ||||||||||||
| M1 a,b | 40.508 | C15H12O7 | [M − H]− | 303.0502 | 303.0510 | −2.64 | 10 | - | ▲ | ▲ | Level 2 | Taxifolin isomer 1 |
| M2 a,b | 42.883 | C15H12O7 | [M − H]− | 303.0517 | 303.0510 | 2.31 | 10 | ▲ | ▲ | ▲ | Level 2 | Taxifolin isomer 2 |
| M3 b | 21.517 | C15H12O10S | [M − H]− | 383.0080 | 383.0078 | 0.52 | 10 | ▲ | - | - | Level 2 | Taxifolin sulphate 1 |
| M4 b | 31.242 | C15H12O10S | [M − H]− | 383.0089 | 383.0078 | 2.87 | 10 | ▲ | - | - | Level 2 | Taxifolin sulphate 2 |
| M5 b | 32.145 | C15H12O10S | [M − H]− | 383.0073 | 383.0078 | −1.31 | 10 | - | ▲ | ▲ | Level 2 | Taxifolin sulphate 3 |
| M6 b | 35.292 | C15H12O10S | [M − H]− | 383.0078 | 383.0078 | 0.00 | 10 | ▲ | - | - | Level 2 | Taxifolin sulphate 4 |
| M7 b | 36.717 | C15H12O10S | [M − H]− | 383.0079 | 383.0078 | 0.26 | 10 | ▲ | ▲ | ▲ | Level 2 | Taxifolin sulphate 5 |
| M8 b | 37.925 | C15H12O10S | [M − H]− | 383.0070 | 383.0078 | −2.09 | 10 | ▲ | ▲ | - | Level 2 | Taxifolin sulphate 6 |
| M9 b | 39.375 | C15H12O10S | [M − H]− | 383.0087 | 383.0078 | 2.35 | 10 | ▲ | - | ▲ | Level 2 | Taxifolin sulphate 7 |
| M10 b | 41.192 | C15H12O10S | [M − H]− | 383.0086 | 383.0078 | 2.09 | 10 | ▲ | ▲ | ▲ | Level 2 | Taxifolin sulphate 8 |
| M11 b | 43.000 | C15H12O10S | [M − H]− | 383.0082 | 383.0078 | 1.04 | 10 | ▲ | ▲ | ▲ | Level 2 | Taxifolin sulphate 9 |
| M12 c | 24.592 | C15H12O13S2 | [M − H]− | 462.9644 | 462.9647 | −0.65 | 10 | ▲ | - | - | Level 3 | Taxifolin disulphate 1 |
| M13 c | 27.458 | C15H12O13S2 | [M − H]− | 462.9670 | 462.9647 | 4.97 | 10 | ▲ | - | - | Level 3 | Taxifolin disulphate 2 |
| M14 c | 31.075 | C15H12O13S2 | [M − H]− | 462.9639 | 462.9647 | −1.73 | 10 | ▲ | - | - | Level 3 | Taxifolin disulphate 3 |
| M15 c | 39.767 | C15H12O13S2 | [M − H]− | 462.9656 | 462.9647 | 1.94 | 10 | ▲ | - | - | Level 3 | Taxifolin disulphate 4 |
| M16 c | 16.252 | C20H19NO13S | [M − H]− | 512.0509 | 512.0504 | 0.98 | 12 | ▲ | - | - | Level 3 | Taxifolin sulphate and pyroglutamic acid conjugate |
| M17 b | 15.408 | C21H20O13 | [M − H]− | 479.0834 | 479.0831 | 0.63 | 12 | ▲ | - | - | Level 2 | Taxifolin glucuronide 1 |
| M18 b | 18.637 | C21H20O13 | [M − H]− | 479.0850 | 479.0831 | 3.97 | 12 | - | ▲ | - | Level 2 | Taxifolin glucuronide 2 |
| M19 b | 20.253 | C21H20O13 | [M − H]− | 479.0847 | 479.0831 | 3.34 | 12 | ▲ | ▲ | - | Level 2 | Taxifolin glucuronide 3 |
| M20 b | 21.370 | C21H20O13 | [M − H]− | 479.0843 | 479.0831 | 2.50 | 12 | ▲ | ▲ | - | Level 2 | Taxifolin glucuronide 4 |
| M21 b | 22.267 | C21H20O13 | [M − H]− | 479.0838 | 479.0831 | 1.46 | 12 | ▲ | ▲ | - | Level 2 | Taxifolin glucuronide 5 |
| M22 b | 22.587 | C21H20O13 | [M − H]− | 479.0847 | 479.0831 | 3.34 | 12 | - | ▲ | - | Level 2 | Taxifolin glucuronide 6 |
| M23 b | 31.862 | C21H20O13 | [M − H]− | 479.0830 | 479.0831 | −0.21 | 12 | ▲ | ▲ | - | Level 2 | Taxifolin glucuronide 7 |
| M24 b | 34.742 | C21H20O13 | [M − H]− | 479.0832 | 479.0831 | 0.21 | 12 | ▲ | - | - | Level 2 | Taxifolin glucuronide 8 |
| M25 b | 37.267 | C21H20O13 | [M − H]− | 479.0834 | 479.0831 | 0.63 | 12 | ▲ | ▲ | - | Level 2 | Taxifolin glucuronide 9 |
| M26 c | 13.888 | C21H20O16S | [M − H]− | 559.0388 | 559.0399 | −1.97 | 12 | - | ▲ | - | Level 3 | Taxifolin glucuronide sulphate 1 |
| M27 c | 16.703 | C21H20O16S | [M − H]− | 559.0423 | 559.0399 | 4.29 | 12 | ▲ | ▲ | - | Level 3 | Taxifolin glucuronide sulphate 2 |
| M28 c | 19.928 | C21H20O16S | [M − H]− | 559.0406 | 559.0399 | 1.25 | 12 | ▲ | ▲ | - | Level 3 | Taxifolin glucuronide sulphate 3 |
| M29 c | 21.812 | C21H20O16S | [M − H]− | 559.0411 | 559.0399 | 2.15 | 12 | - | ▲ | - | Level 3 | Taxifolin glucuronide sulphate 4 |
| M30 c | 23.087 | C21H20O16S | [M − H]− | 559.0418 | 559.0399 | 3.40 | 12 | ▲ | ▲ | - | Level 3 | Taxifolin glucuronide sulphate 5 |
| M31 c | 24.762 | C21H20O16S | [M − H]− | 559.0425 | 559.0399 | 4.65 | 12 | - | ▲ | - | Level 3 | Taxifolin glucuronide sulphate 6 |
| M32 c | 25.797 | C21H20O16S | [M − H]− | 559.0411 | 559.0399 | 2.86 | 12 | - | - | - | Level 3 | Taxifolin glucuronide sulphate 7 |
| Metabolites having the aglycone of methyl taxifolin (M33–M69) | ||||||||||||
| M33 b,d | 50.292 | C16H14O7 | [M − H]− | 317.0675 | 317.0667 | 2.52 | 10 | ▲ | ▲ | ▲ | Level 2 | 3′-O-Methyltaxifolin |
| M34 b,d | 51.350 | C16H14O7 | [M − H]− | 317.0673 | 317.0667 | 1.89 | 10 | ▲ | ▲ | ▲ | Level 2 | 4′-O-Methyltaxifolin |
| M35 b,d | 52.875 | C16H14O7 | [M − H]− | 317.0667 | 317.0667 | 0.00 | 10 | ▲ | ▲ | ▲ | Level 2 | 7-O-Methyltaxifolin |
| M36 b,d | 53.592 | C16H14O7 | [M − H]− | 317.0660 | 317.0667 | −2.21 | 10 | ▲ | - | - | Level 2 | 3-O-Methyltaxifolin |
| M37 c | 28.575 | C16H14O10S | [M − H]− | 397.0243 | 397.0235 | 2.01 | 10 | ▲ | - | - | Level 3 | Methyl taxifolin sulphate 1 |
| M38 c | 33.942 | C16H14O10S | [M − H]− | 397.0240 | 397.0235 | 1.26 | 10 | ▲ | ▲ | - | Level 3 | Methyl taxifolin sulphate 2 |
| M39 c | 34.420 | C16H14O10S | [M − H]− | 397.0247 | 397.0235 | 3.02 | 10 | - | ▲ | - | Level 3 | Methyl taxifolin sulphate 3 |
| M40 c | 35.858 | C16H14O10S | [M − H]− | 397.0253 | 397.0235 | 4.53 | 10 | ▲ | - | - | Level 3 | Methyl taxifolin sulphate 4 |
| M41 c | 38.092 | C16H14O10S | [M − H]− | 397.0241 | 397.0235 | 1.51 | 10 | ▲ | - | - | Level 3 | Methyl taxifolin sulphate 5 |
| M42 c | 40.283 | C16H14O10S | [M − H]− | 397.0233 | 397.0235 | −0.50 | 10 | ▲ | ▲ | ▲ | Level 3 | Methyl taxifolin sulphate 6 |
| M43 c | 41.817 | C16H14O10S | [M − H]− | 397.0241 | 397.0235 | 1.51 | 10 | ▲ | ▲ | ▲ | Level 3 | Methyl taxifolin sulphate 7 |
| M44 c | 42.717 | C16H14O10S | [M − H]− | 397.0230 | 397.0235 | −1.26 | 10 | ▲ | - | - | Level 3 | Methyl taxifolin sulphate 8 |
| M45 c | 43.600 | C16H14O10S | [M − H]− | 397.0235 | 397.0235 | 0.00 | 10 | ▲ | ▲ | - | Level 3 | Methyl taxifolin sulphate 9 |
| M46 c | 45.558 | C16H14O10S | [M − H]− | 397.0238 | 397.0235 | 0.76 | 10 | ▲ | ▲ | ▲ | Level 3 | Methyl taxifolin sulphate 10 |
| M47 b | 23.520 | C22H22O13 | [M − H]− | 493.0973 | 493.0988 | −3.04 | 12 | - | ▲ | - | Level 2 | Methyl taxifolin glucuronide 1 |
| M48 b | 25.212 | C22H22O13 | [M − H]− | 493.1012 | 493.0988 | 4.87 | 12 | - | ▲ | - | Level 2 | Methyl taxifolin glucuronide 2 |
| M49 b | 26.687 | C22H22O13 | [M − H]− | 493.1012 | 493.0988 | 4.87 | 12 | - | ▲ | - | Level 2 | Methyl taxifolin glucuronide 3 |
| M50 b | 30.383 | C22H22O13 | [M − H]− | 493.1012 | 493.0988 | 4.87 | 12 | ▲ | ▲ | - | Level 2 | Methyl taxifolin glucuronide 4 |
| M51 b | 33.395 | C22H22O13 | [M − H]− | 493.1007 | 493.0988 | 3.85 | 12 | - | ▲ | - | Level 2 | Methyl taxifolin glucuronide 5 |
| M52 b | 35.692 | C22H22O13 | [M − H]− | 493.0998 | 493.0988 | 2.03 | 12 | ▲ | ▲ | - | Level 2 | Methyl taxifolin glucuronide 6 |
| M53 b | 36.025 | C22H22O13 | [M − H]− | 493.1004 | 493.0988 | 3.24 | 12 | ▲ | - | - | Level 2 | Methyl taxifolin glucuronide 7 |
| M54 b | 37.600 | C22H22O13 | [M − H]− | 493.0998 | 493.0988 | 2.03 | 12 | ▲ | - | - | Level 2 | Methyl taxifolin glucuronide 8 |
| M55 b | 42.375 | C22H22O13 | [M − H]− | 493.1008 | 493.0988 | 4.06 | 12 | ▲ | - | - | Level 2 | Methyl taxifolin glucuronide 9 |
| M56 | 34.742 | C22H22O16S | [M − H]− | 573.0560 | 573.0556 | 0.70 | 12 | ▲ | - | - | Level 2 | Methyl taxifolin glucuronide sulphate 1 |
| M57 | 37.158 | C22H22O16S | [M − H]− | 573.0533 | 573.0556 | −4.01 | 12 | ▲ | - | - | Level 2 | Methyl taxifolin glucuronide sulphate 2 |
| M58 c | 16.490 | C21H21NO10 | [M − H]− | 446.1107 | 446.1093 | 3.14 | 12 | ▲ | - | - | Level 3 | Methyl taxifolin pyroglutamic acid conjugate 1 |
| M59 c | 18.483 | C21H21NO10 | [M − H]− | 446.1086 | 446.1093 | −1.57 | 12 | ▲ | - | - | Level 3 | Methyl taxifolin pyroglutamic acid conjugate 2 |
| M60 c | 37.848 | C16H14O11S | [M − H]− | 413.0200 | 413.0184 | 3.87 | 10 | - | ▲ | - | Level 3 | Hydroxylated methyl taxifolin sulphate 1 |
| M61 c | 41.943 | C16H14O11S | [M − H]− | 413.0175 | 413.0184 | −2.18 | 10 | - | ▲ | - | Level 3 | Hydroxylated methyl taxifolin sulphate 2 |
| M62 c | 42.375 | C16H14O11S | [M − H]− | 413.0198 | 413.0184 | 3.39 | 10 | ▲ | - | - | Level 3 | Hydroxylated methyl taxifolin sulphate 3 |
| M63 c | 42.660 | C16H14O11S | [M − H]− | 413.0191 | 413.0184 | 1.69 | 10 | - | ▲ | - | Level 3 | Hydroxylated methyl taxifolin sulphate 4 |
| M64 c | 55.808 | C16H14O9 | [M − H]− | 349.0580 | 349.0565 | 4.30 | 10 | ▲ | - | ▲ | Level 3 | Methylated and dihydroxylated taxifolin 1 |
| M65 c | 56.608 | C16H14O9 | [M − H]− | 349.0551 | 349.0565 | −4.01 | 10 | - | - | - | Level 3 | Methylated and dihydroxylated taxifolin 2 |
| M66 c | 17.170 | C22H22O15 | [M − H]− | 525.0865 | 525.0886 | −4.00 | 12 | - | ▲ | - | Level 3 | Methylated and dihydroxylated taxifolin glucuronide 1 |
| M67 c | 17.887 | C22H22O15 | [M − H]− | 525.0908 | 525.0886 | 4.19 | 12 | - | ▲ | - | Level 3 | Methylated and dihydroxylated taxifolin glucuronide 2 |
| M68 c | 18.637 | C22H22O15 | [M − H]− | 525.0890 | 525.0886 | 0.76 | 12 | - | ▲ | - | Level 3 | Methylated and dihydroxylated taxifolin glucuronide 3 |
| M69 c | 19.178 | C22H22O15 | [M − H]− | 525.0911 | 525.0886 | 4.76 | 12 | - | ▲ | - | Level 3 | Methylated and dihydroxylated taxifolin glucuronide 4 |
| Metabolites having the aglycone of quercetin(M70–M103); eight bioactive metabolites | ||||||||||||
| M70 a,d | 58.150 | C15H10O7 | [M − H]− | 301.0350 | 301.0354 | −1.33 | 11 | ▲ | - | ▲ | Level 2 | Quercetin |
| M71 d | 51.583 | C15H10O10S | [M − H]− | 380.9933 | 380.9922 | 0.26 | 11 | ▲ | - | - | Level 2 | Quercetin-5 -O-sulphate |
| M72 d | 52.647 | C15H10O10S | [M − H]− | 380.9932 | 380.9922 | 2.89 | 11 | - | - | - | Level 2 | Quercetin-7-O-sulphate |
| M73 a,d | 56.300 | C15H10O10S | [M − H]− | 380.9922 | 380.9922 | 0.00 | 11 | ▲ | - | ▲ | Level 2 | Quercetin-4′-O-sulphate |
| M74 a,d | 57.033 | C15H10O10S | [M − H]− | 380.9932 | 380.9922 | 2.62 | 11 | ▲ | - | ▲ | Level 2 | Quercetin-3′-O-sulphate |
| M75 a,d | 58.173 | C15H10O10S | [M − H]− | 380.9937 | 380.9922 | 3.94 | 11 | - | - | - | Level 2 | Quercetin-3-O-sulphate |
| M76 a | 37.542 | C21H18O13 | [M − H]− | 477.0688 | 477.0675 | 2.72 | 13 | ▲ | - | - | Level 2 | Quercetin glucuronide |
| M77 | 40.727 | C21H18O16S | [M − H]− | 557.0252 | 557.0243 | 1.62 | 13 | - | ▲ | - | Level 2 | Quercetin glucuronide sulphate 1 |
| M78 | 41.068 | C21H18O16S | [M − H]− | 557.0268 | 557.0243 | 4.49 | 13 | - | ▲ | - | Level 2 | Quercetin glucuronide sulphate 2 |
| M79 | 41.443 | C21H18O16S | [M − H]− | 557.0269 | 557.0243 | 4.67 | 13 | - | ▲ | - | Level 2 | Quercetin glucuronide sulphate 3 |
| M80 a,d | 65.417 | C16H12O7 | [M − H]− | 315.0503 | 315.0510 | −2.22 | 11 | ▲ | - | ▲ | Level 2 | Isorhamnetin |
| M81 d | 48.633 | C16H12O10S | [M − H]− | 395.0081 | 395.0078 | 0.76 | 11 | ▲ | ▲ | - | Level 2 | Isorhamnetin-5-O-sulphate |
| M82 d | 56.917 | C16H12O10S | [M − H]− | 395.0082 | 395.0078 | 1.01 | 11 | ▲ | - | ▲ | Level 2 | Isorhamnetin-7-O-sulphate |
| M83 a,d | 58.042 | C16H12O10S | [M − H]− | 395.0085 | 395.0078 | 1.77 | 11 | ▲ | ▲ | ▲ | Level 2 | Isorhamnetin-3-O-sulphate |
| M84 d | 58.922 | C16H12O10S | [M − H]− | 395.0082 | 395.0078 | 1.01 | 11 | - | ▲ | - | Level 2 | Isorhamnetin-4′-O-sulphate |
| M85 a | 48.308 | C16H12O13S2 | [M − H]− | 474.9658 | 474.9647 | 2.32 | 11 | ▲ | ▲ | - | Level 2 | Isorhamnetin disulphate |
| M86 d | 49.212 | C22H20O13 | [M − H]− | 491.0852 | 491.0831 | 4.28 | 13 | - | ▲ | - | Level 2 | Isorhamnetin-4′-O-glucuronide |
| M87 d | 50.428 | C22H20O13 | [M − H]− | 491.0836 | 491.0831 | 1.02 | 13 | - | ▲ | - | Level 2 | Isorhamnetin-7-O-glucuronide |
| M88 | 40.143 | C22H20O16S | [M − H]− | 571.0381 | 571.0399 | −3.15 | 13 | - | ▲ | - | Level 2 | Isorhamnetin glucuronide sulphate 1 |
| M89 | 41.118 | C22H20O16S | [M − H]− | 571.0413 | 571.0399 | 2.45 | 13 | - | ▲ | - | Level 2 | Isorhamnetin glucuronide sulphate 2 |
| M90 | 44.673 | C22H20O16S | [M − H]− | 571.0395 | 571.0399 | −0.70 | 13 | - | ▲ | - | Level 2 | Isorhamnetin glucuronide sulphate 3 |
| M91 | 45.392 | C22H20O16S | [M − H]− | 571.0419 | 571.0399 | 3.50 | 13 | - | - | - | Level 2 | Isorhamnetin glucuronide sulphate 4 |
| M92 | 27.987 | C15H10O11S | [M − H]− | 396.9882 | 396.9871 | 2.77 | 11 | - | ▲ | - | Level 2 | Hydroxylated quercetin sulphate 1 |
| M93 | 28.487 | C15H10O11S | [M − H]− | 396.9868 | 396.9871 | −0.76 | 11 | - | ▲ | - | Level 2 | Hydroxylated quercetin sulphate 2 |
| M94 | 29.028 | C15H10O11S | [M − H]− | 396.9876 | 396.9871 | 1.26 | 11 | - | ▲ | - | Level 2 | Hydroxylated quercetin sulphate 3 |
| M95 | 15.930 | C21H18O14 | [M − H]− | 493.0642 | 493.0624 | 3.65 | 13 | - | ▲ | - | Level 2 | Hydroxylated quercetin glucuronide 1 |
| M96 | 17.720 | C21H18O14 | [M − H]− | 493.0601 | 493.0624 | −4.66 | 13 | - | ▲ | - | Level 2 | Hydroxylated quercetin glucuronide 2 |
| M97 | 39.160 | C16H12O11S | [M − H]− | 411.0022 | 411.0028 | −1.46 | 11 | - | ▲ | - | Level 2 | Hydroxylated isorhamnetin sulphate 1 |
| M98 | 39.710 | C16H12O11S | [M − H]− | 411.0043 | 411.0028 | 3.65 | 11 | - | ▲ | - | Level 2 | Hydroxylated isorhamnetin sulphate 2 |
| M99 | 40.193 | C16H12O11S | [M − H]− | 411.0039 | 411.0028 | 2.68 | 11 | - | ▲ | - | Level 2 | Hydroxylated isorhamnetin sulphate 3 |
| M100 | 59.017 | C16H12O11S | [M − H]− | 411.0030 | 411.0028 | 0.49 | 11 | ▲ | ▲ | - | Level 2 | Hydroxylated isorhamnetin sulphate 4 |
| M101 | 25.103 | C22H20O14 | [M − H]− | 507.0790 | 507.0780 | 1.97 | 13 | - | ▲ | - | Level 2 | Hydroxylated isorhamnetin glucuronide 1 |
| M102 | 25.728 | C22H20O14 | [M − H]− | 507.0758 | 507.0780 | −4.34 | 13 | - | ▲ | - | Level 2 | Hydroxylated isorhamnetin glucuronide 2 |
| M103 | 26.570 | C22H20O14 | [M − H]− | 507.0805 | 507.0780 | 4.93 | 13 | - | ▲ | - | Level 2 | Hydroxylated isorhamnetin glucuronide 3 |
| Metabolites having the aglycone of dehydroxylated taxifolin (M104–M112); two bioactive metabolites | ||||||||||||
| M104 a,b,d | 40.733 | C15H12O6 | [M − H]− | 287.0557 | 287.0561 | −1.39 | 10 | - | - | ▲ | Level 2 | Eriodictyol |
| M105 a,b,d | 49.442 | C15H12O6 | [M − H]− | 287.0555 | 287.0561 | −2.09 | 10 | ▲ | - | ▲ | Level 2 | Dihydrokaempferol |
| M106 d | 37.325 | C15H12O9S | [M − H]− | 367.0128 | 367.0129 | −0.27 | 10 | ▲ | - | - | Level 2 | Eriodictyol-7-O-sulphate |
| M107 d | 37.708 | C15H12O9S | [M − H]− | 367.0144 | 367.0129 | 4.09 | 10 | ▲ | - | ▲ | Level 2 | Dihydrokaempferol-7-O-sulphate |
| M108 d | 38.200 | C15H12O9S | [M − H]− | 367.0144 | 367.0129 | 4.09 | 10 | ▲ | - | ▲ | Level 2 | Eriodictyol-3′-O-sulphate |
| M109 d | 40.383 | C15H12O9S | [M − H]− | 367.0123 | 367.0129 | −1.63 | 10 | - | - | - | Level 2 | Dihydrokaempferol-4′-O-sulphate |
| M110 | 28.045 | C21H20O12 | [M − H]− | 463.0907 | 463.0882 | 5.40 | 12 | - | ▲ | - | Level 2 | Dehydroxylated taxifolin glucuronide 1 |
| M111 | 28.753 | C21H20O12 | [M − H]− | 463.0856 | 463.0882 | −5.61 | 12 | - | ▲ | - | Level 2 | Dehydroxylated taxifolin glucuronide 2 |
| M112 d | 28.970 | C21H20O12 | [M − H]− | 463.0888 | 463.0882 | 1.30 | 12 | - | ▲ | - | Level 2 | Dihydrokaempferol-4′-O-glucuronide |
| Metabolites formed through dehydration and glucuronidation (M113–M116); one bioactive metabolite | ||||||||||||
| M113 a,d | 16.017 | C21H18O12 | [M + NH2]− | 478.1007 | 478.0991 | 3.35 | 13 | ▲ | - | - | Level 2 | Luteolin-7-O-glucuronide |
| M114 d | 16.525 | C21H18O12 | [M + NH2]− | 478.1007 | 478.0991 | 3.35 | 13 | ▲ | - | - | Level 2 | Luteolin-3′/4′-O-glucuronide |
| M115 d | 17.425 | C21H18O12 | [M + NH2]− | 478.1014 | 478.0991 | 4.81 | 13 | ▲ | - | - | Level 2 | Luteolin-3′/4′-O-glucuronide |
| M116 | 23.625 | C22H20O12 | [M + NH2]− | 492.1160 | 492.1147 | 2.64 | 13 | ▲ | - | - | Level 2 | Methyl luteolin glucuronide |
| Metabolites having the aglycone of hydrogenated taxifolin (M117–M121) | ||||||||||||
| M117 | 43.883 | C15H14O7 | [M − H]− | 305.0652 | 305.0667 | −4.92 | 9 | ▲ | - | ▲ | Level 2 | Hydrogenated taxifolin |
| M118 | 52.325 | C16H16O7 | [M − H]− | 319.0813 | 319.0823 | −3.13 | 9 | ▲ | - | ▲ | Level 2 | Hydrogenated methyl taxifolin |
| M119 | 38.567 | C15H14O10S | [M − H]− | 385.0224 | 385.0235 | −2.86 | 9 | - | - | ▲ | Level 2 | Hydrogenated taxifolin sulphate 1 |
| M120 | 43.433 | C15H14O10S | [M − H]− | 385.0224 | 385.0235 | −2.86 | 9 | - | - | ▲ | Level 2 | Hydrogenated taxifolin sulphate 2 |
| M121 | 45.442 | C15H14O10S | [M − H]− | 385.0227 | 385.0235 | −2.08 | 9 | ▲ | - | ▲ | Level 2 | Hydrogenated taxifolin sulphate 3 |
| Phenolic acid metabolites through ring cleavage (M122–M159); four bioactive metabolites | ||||||||||||
| M122 a,b,d | 35.317 | C9H10O3 | [M − H]− | 165.0555 | 165.0557 | −1.21 | 5 | - | - | ▲ | Level 2 | 3/4-Hydroxyphenylpropionic acid |
| M123 a,d | 35.917 | C9H10O3 | [M − H]− | 165.0559 | 165.0557 | 1.21 | 5 | ▲ | - | ▲ | Level 2 | 3/4-Hydroxyphenylpropionic acid |
| M124 d | 21.712 | C9H10O6S | [M − H]− | 245.0132 | 245.0125 | 2.86 | 5 | ▲ | - | - | Level 2 | 4-Hydroxyphenylpropionic acid sulphate |
| M125 d | 23.683 | C9H10O6S | [M − H]− | 245.0133 | 245.0125 | 3.27 | 5 | ▲ | ▲ | - | Level 2 | 3-Hydroxyphenylpropionic acid sulphate |
| M126 d | 23.787 | C15H18O9 | [M − H]− | 341.0866 | 341.0878 | −1.76 | 7 | ▲ | - | - | Level 2 | 3/4-Hydroxyphenylpropionic acid glucuronide |
| M127 d | 24.078 | C15H18O9 | [M − H]− | 341.0891 | 341.0878 | 3.81 | 7 | ▲ | - | - | Level 2 | 3/4-Hydroxyphenylpropionic acid glucuronide |
| M128 d | 22.325 | C9H8O6S | [M − H]− | 242.9969 | 242.9969 | 0.00 | 6 | ▲ | - | - | Level 2 | p/m-Coumaric acid sulphate |
| M129 d | 25.758 | C9H8O6S | [M − H]− | 242.9972 | 242.9969 | 1.23 | 6 | ▲ | ▲ | - | Level 2 | p/m-Coumaric acid sulphate |
| M130 d | 27.067 | C9H8O6S | [M − H]− | 242.9971 | 242.9969 | 0.82 | 6 | ▲ | - | - | Level 2 | p/m-Coumaric acid sulphate |
| M131 a,b,d | 16.490 | C8H8O4 | [M − H]− | 167.0349 | 167.0350 | −0.60 | 5 | - | - | ▲ | Level 2 | Dihydroxyphenylacetic acid |
| M132 d | 16.258 | C8H8O7S | [M − H]− | 246.9927 | 246.9918 | 3.64 | 5 | ▲ | - | ▲ | Level 2 | Dihydroxyphenylacetic acid sulfae 1 |
| M133 d | 15.800 | C8H8O7S | [M − H]− | 246.9927 | 246.9918 | 3.64 | 5 | - | - | ▲ | Level 2 | Dihydroxyphenylacetic acid sulfae 2 |
| M134 d | 16.933 | C8H8O7S | [M − H]− | 246.9920 | 246.9918 | 0.81 | 5 | ▲ | - | ▲ | Level 2 | Dihydroxyphenylacetic acid sulfae 3 |
| M135 d | 18.108 | C9H10O7S | [M − H]− | 261.0073 | 261.0074 | −0.38 | 5 | ▲ | - | - | Level 2 | Homovanillic acid sulphate |
| M136 d | 22.508 | C9H10O4 | [M − H]− | 181.0504 | 181.0506 | −1.10 | 5 | - | - | ▲ | Level 2 | Dihydrocaffeic acid |
| M137 d | 20.033 | C9H10O7S | [M − H]− | 261.0082 | 261.0074 | 3.07 | 5 | ▲ | - | - | Level 2 | Dihydrocaffeic acid sulphate 1 |
| M138 d | 20.942 | C9H10O7S | [M − H]− | 261.0084 | 261.0074 | 3.83 | 5 | ▲ | - | - | Level 2 | Dihydrocaffeic acid sulphate 2 |
| M139 d | 13.108 | C11H13NO5 | [M − H]− | 238.0720 | 238.0721 | −0.42 | 6 | - | - | ▲ | Level 2 | Caffeic acid acetamide 1 |
| M140 d | 13.592 | C11H13NO5 | [M − H]− | 238.0724 | 238.0721 | 1.26 | 6 | - | - | ▲ | Level 2 | Caffeic acid acetamide 2 |
| M141 d | 13.858 | C11H13NO5 | [M − H]− | 238.0728 | 238.0721 | 2.94 | 6 | - | - | ▲ | Level 2 | Caffeic acid acetamide 3 |
| M142 d | 11.692 | C9H10O5 | [M − H]− | 197.0461 | 197.0455 | 3.04 | 5 | ▲ | - | ▲ | Level 2 | 3-(3,4-Dihydroxyphenyl)-3-hydroxypropanoic acid |
| M143 d | 12.658 | C9H10O5 | [M − H]− | 197.0456 | 197.0455 | 0.51 | 5 | ▲ | - | ▲ | Level 2 | 3-(3,4-Dihydroxyphenyl)-2-hydroxypropanoic acid |
| M144 d | 12.700 | C9H10O8S | [M − H]− | 277.0024 | 277.0024 | 0.00 | 5 | ▲ | - | ▲ | Level 2 | Caffeic acid hydrate sulphate 1 |
| M145 d | 13.433 | C9H10O8S | [M − H]− | 277.0025 | 277.0024 | 0.36 | 5 | ▲ | - | ▲ | Level 2 | Caffeic acid hydrate sulphate 2 |
| M146 d | 22.667 | C10H12O7S | [M − H]− | 275.0236 | 275.0231 | 1.82 | 5 | ▲ | - | - | Level 2 | Dihydrogen ferulic acid sulphate |
| M147 d | 15.810 | C10H12O8S | [M − H]− | 291.0174 | 291.0180 | 3.78 | 5 | ▲ | - | - | Level 2 | Ferulic acid hydrate sulphate 1 |
| M148 d | 16.233 | C10H12O8S | [M − H]− | 291.0184 | 291.0180 | 1.37 | 5 | ▲ | - | - | Level 2 | Ferulic acid hydrate sulphate 2 |
| M149 | 25.208 | C7H8O5S | [M − H]− | 203.0021 | 203.0020 | 0.49 | 4 | ▲ | - | - | Level 2 | Hydroxybenzyl alcohol sulphate |
| M150 d | 29.025 | C13H16O8 | [M − H]− | 299.0773 | 299.0772 | 0.33 | 6 | ▲ | ▲ | - | Level 2 | Hydroxybenzyl alcohol glucuronide 1 |
| M151 d | 29.717 | C13H16O8 | [M − H]− | 299.0771 | 299.0772 | −0.33 | 6 | ▲ | ▲ | - | Level 2 | Hydroxybenzyl alcohol glucuronide 2 |
| M152 c,d | 18.795 | C13H16O11S | [M − H]− | 379.0336 | 379.0341 | −1.32 | 6 | - | ▲ | - | Level 3 | Hydroxybenzyl alcohol glucuronide sulphate 1 |
| M153 c,d | 21.095 | C13H16O11S | [M − H]− | 379.0337 | 379.0341 | −1.06 | 6 | - | ▲ | - | Level 3 | Hydroxybenzyl alcohol glucuronide sulphate 2 |
| M154 c,d | 33.083 | C8H10O5S | [M − H]− | 217.0168 | 217.0176 | −3.69 | 4 | ▲ | - | - | Level 3 | Methyl hydroxybenzyl alcohol sulphate 1 |
| M155 c,d | 34.625 | C8H10O5S | [M − H]− | 217.0181 | 217.0176 | 2.30 | 4 | ▲ | - | - | Level 3 | Methyl hydroxybenzyl alcohol sulphate 2 |
| M156 d | 17.512 | C7H6O6S | [M − H]− | 216.9822 | 216.9812 | 4.61 | 5 | - | ▲ | - | Level 2 | 3/4-Hydroxy benzoic acid sulphate |
| M157 d | 17.937 | C7H6O6S | [M − H]− | 216.9810 | 216.9812 | −0.92 | 5 | - | ▲ | - | Level 2 | 3/4-Hydroxy benzoic acid sulphate |
| M158 d | 30.987 | C8H8O7S | [M − H]− | 246.9914 | 246.9918 | −1.62 | 5 | - | ▲ | - | Level 2 | Vanillic acid sulphate |
| M159 d | 31.978 | C8H8O7S | [M − H]− | 246.9909 | 246.9918 | −3.64 | 5 | - | ▲ | - | Level 2 | Isovanillic acid sulphate |
| Metabolites formed through polymerization(M160–M191) | ||||||||||||
| M160 | 61.342 | C31H24O13 | [M − H]− | 603.1151 | 603.1144 | 1.16 | 20 | ▲ | - | - | Level 2 | Dimer of taxiflolin and dehydroxylated methyl taxifolin |
| M161 c | 55.533 | C31H24O14 | [M − H]− | 619.1063 | 619.1093 | −4.85 | 20 | ▲ | - | - | Level 3 | Dimer of taxiflolin and methyl taxifolin 1 |
| M162 c | 60.600 | C31H24O14 | [M − H]− | 619.1090 | 619.1093 | −0.48 | 20 | ▲ | - | - | Level 3 | Dimer of taxiflolin and methyl taxifolin 2 |
| M163 c | 64.608 | C32H26O14 | [M − H]− | 633.1249 | 633.1250 | −0.16 | 20 | ▲ | - | - | Level 3 | Dimer of taxiflolin and dimethyl taxifolin |
| M164 c | 56.025 | C31H24O17S | [M − H]− | 699.0699 | 699.0661 | 5.44 | 20 | ▲ | - | - | Level 3 | Dimer of taxiflolin and methyl taxifolin sulphate 1 |
| M165 c | 56.750 | C31H24O17S | [M − H]− | 699.0671 | 699.0661 | 1.43 | 20 | ▲ | - | - | Level 3 | Dimer of taxiflolin and methyl taxifolin sulphate 2 |
| M166 c | 60.817 | C31H24O17S | [M − H]− | 699.0678 | 699.0661 | 2.43 | 20 | ▲ | ▲ | - | Level 3 | Dimer of taxiflolin and methyl taxifolin sulphate 3 |
| M167 c | 59.725 | C32H26O17S | [M − H]− | 713.0844 | 713.0818 | 3.65 | 20 | ▲ | - | - | Level 3 | Dimer of taxiflolin and dimethyl taxifolin sulphate 1 |
| M168 c | 60.167 | C32H26O17S | [M − H]− | 713.0839 | 713.0818 | 2.94 | 20 | ▲ | - | - | Level 3 | Dimer of taxiflolin and dimethyl taxifolin sulphate 2 |
| M169 c | 64.125 | C32H26O17S | [M − H]− | 713.0843 | 713.0818 | 3.51 | 20 | ▲ | ▲ | - | Level 3 | Dimer of taxiflolin and dimethyl taxifolin sulphate 3 |
| M170 c | 60.650 | C32H26O13 | [M − H]− | 617.1291 | 617.1301 | −1.62 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dehydroxylated methyl taxifolin 1 |
| M171 c | 64.400 | C32H26O13 | [M − H]− | 617.1311 | 617.1301 | 1.62 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dehydroxylated methyl taxifolin 2 |
| M172 c | 64.925 | C32H26O13 | [M − H]− | 617.1299 | 617.1301 | −0.32 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dehydroxylated methyl taxifolin 3 |
| M173 c | 65.142 | C32H24O14 | [M − H]− | 631.1093 | 631.1093 | 0.00 | 21 | ▲ | - | - | Level 3 | Dimer of methyl quercetin and methyl taxifolin 1 |
| M174 c | 66.142 | C32H24O14 | [M − H]− | 631.1088 | 631.1093 | −0.79 | 21 | ▲ | - | - | Level 3 | Dimer of methyl quercetin and methyl taxifolin 2 |
| M175 c | 68.517 | C32H24O14 | [M − H]− | 631.1106 | 631.1093 | 2.06 | 21 | ▲ | - | - | Level 3 | Dimer of methyl quercetin and methyl taxifolin 3 |
| M176 c | 69.230 | C32H24O14 | [M − H]− | 631.1105 | 631.1093 | 1.90 | 21 | ▲ | - | - | Level 3 | Dimer of methyl quercetin and methyl taxifolin 4 |
| M177 c | 64.550 | C33H28O13 | [M − H]− | 631.1435 | 631.1457 | −3.49 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dehydroxylated dimethyl taxifolin 1 |
| M178 c | 67.408 | C33H28O13 | [M − H]− | 631.1482 | 631.1457 | 3.96 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dehydroxylated dimethyl taxifolin 2 |
| M179 c | 67.633 | C33H28O13 | [M − H]− | 631.1488 | 631.1457 | 4.91 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dehydroxylated dimethyl taxifolin 3 |
| M180 c | 59.138 | C32H26O14 | [M − H]− | 633.1257 | 633.1250 | 1.11 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and methyl taxifolin 1 |
| M181 c | 63.783 | C32H26O14 | [M − H]− | 633.1252 | 633.1250 | 0.32 | 20 | ▲ | ▲ | - | Level 3 | Dimer of methyl taxiflolin and methyl taxifolin 2 |
| M182 c | 69.755 | C33H26O14 | [M − H]− | 645.1243 | 645.1250 | −1.09 | 21 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dimethyl quercetin 1 |
| M183 c | 71.097 | C33H26O14 | [M − H]− | 645.1252 | 645.1250 | 0.31 | 21 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dimethyl quercetin 2 |
| M184 c | 62.067 | C33H28O14 | [M − H]− | 647.1432 | 647.1406 | 4.02 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dimethyl taxifolin 1 |
| M185 c | 62.600 | C33H28O14 | [M − H]− | 647.1420 | 647.1406 | 2.16 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dimethyl taxifolin 2 |
| M186 c | 62.917 | C33H28O14 | [M − H]− | 647.1419 | 647.1406 | 2.01 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dimethyl taxifolin 3 |
| M187 c | 63.183 | C33H28O14 | [M − H]− | 647.1406 | 647.1406 | 0.00 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dimethyl taxifolin 4 |
| M188 c | 66.483 | C33H28O14 | [M − H]− | 647.1434 | 647.1406 | 4.33 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dimethyl taxifolin 5 |
| M189 c | 66.983 | C33H28O14 | [M − H]− | 647.1405 | 647.1406 | −0.15 | 20 | ▲ | ▲ | - | Level 3 | Dimer of methyl taxiflolin and dimethyl taxifolin 6 |
| M190 c | 70.430 | C33H28O14 | [M − H]− | 647.1421 | 647.1406 | 2.32 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dimethyl taxifolin 7 |
| M191 c | 63.958 | C32H26O16S | [M − H]− | 697.0891 | 697.0869 | 3.16 | 20 | ▲ | - | - | Level 3 | Dimer of methyl taxiflolin and dehydroxylated methyl taxifolin sulphate |
| Metabolic Reaction | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Amount | Phase I | Phase II | ||||||||||||
| −H2O | −OH | +OH | −2H | +2H | RC | I | P | CH3 | +SO3H | +GlcUA | +AA c | +AM c | |||
| M1, M2 | 2 | ▲ | |||||||||||||
| M3–M11 | 9 | ▲ | |||||||||||||
| M12–M15 | 4 | ▲ a | |||||||||||||
| M16 | 1 | ▲ | ▲ | ||||||||||||
| M17–M25 | 9 | ▲ | |||||||||||||
| M26–M32 | 7 | ▲ | ▲ | ▲ | |||||||||||
| M33–M36 | 4 | ▲ | |||||||||||||
| M37–M46 | 10 | ▲ | ▲ | ||||||||||||
| M47–M55 | 9 | ▲ | ▲ | ||||||||||||
| M56, M57 | 2 | ▲ | ▲ | ▲ | |||||||||||
| M58, M59 | 2 | ▲ | ▲ | ||||||||||||
| M60–M63 | 4 | ▲ | ▲ | ▲ | |||||||||||
| M64, M65 | 2 | ▲ a | ▲ | ||||||||||||
| M66–M69 | 4 | ▲ a | ▲ | ▲ | |||||||||||
| M70 | 1 | ▲ | |||||||||||||
| M71–M75 | 5 | ▲ | ▲ | ||||||||||||
| M76 | 1 | ▲ | ▲ | ||||||||||||
| M77–M79 | 3 | ▲ | ▲ | ||||||||||||
| M80 | 1 | ▲ | ▲ | ▲ | |||||||||||
| M81–M84 | 4 | ▲ | ▲ | ▲ | |||||||||||
| M85 | 1 | ▲ | ▲ | ▲ a | ▲ | ||||||||||
| M86, M87 | 2 | ▲ | ▲ | ▲ | |||||||||||
| M88–M91 | 4 | ▲ | ▲ | ▲ | ▲ | ||||||||||
| M92–M94 | 3 | ▲ | ▲ | ▲ | |||||||||||
| M95, M96 | 2 | ▲ | ▲ | ▲ | |||||||||||
| M97–M100 | 4 | ▲ | ▲ | ▲ | ▲ | ||||||||||
| M101–M103 | 3 | ▲ | ▲ | ▲ | ▲ | ||||||||||
| M104, M105 | 2 | ▲ | |||||||||||||
| M106–M109 | 4 | ▲ | ▲ | ||||||||||||
| M110–M112 | 3 | ▲ | ▲ | ||||||||||||
| M113–M115 | 3 | ▲ | ▲ | ||||||||||||
| M116 | 1 | ▲ | ▲ | ▲ | |||||||||||
| M117 | 1 | ▲ | |||||||||||||
| M118 | 1 | ▲ | ▲ | ||||||||||||
| M119–M121 | 3 | ▲ | ▲ | ||||||||||||
| M122, M123, M131, M136, M142, M143 | 6 | ▲ | ▲ | ||||||||||||
| M124, M125, M128–M130, M132–M135,M137, M138, 144-M149, M154–M159 | 23 | ▲ | ▲ | ||||||||||||
| M126, M127, M150, M151 | 4 | ▲ | ▲ | ||||||||||||
| M139–M141 | 3 | ▲ | ▲ | ||||||||||||
| M152, M153 | 2 | ▲ | ▲ | ▲ | |||||||||||
| M160 | 1 | ▲ | ▲ | ▲ | |||||||||||
| M161–M162 | 2 | ▲ | ▲ | ||||||||||||
| M163, M180, M181 | 3 | ▲ | ▲ a | ||||||||||||
| M164–M166 | 3 | ▲ | ▲ | ▲ | |||||||||||
| M167–M169 | 3 | ▲ | ▲ a | ▲ | |||||||||||
| M170–M172 | 3 | ▲ | ▲ | ▲ a | |||||||||||
| M173–M176 | 4 | ▲ | ▲ | ▲ a | |||||||||||
| M177–M179 | 3 | ▲ | ▲ | ▲ b | |||||||||||
| M182, M183 | 2 | ▲ | ▲ | ▲ b | |||||||||||
| M184–M190 | 7 | ▲ | ▲ b | ||||||||||||
| M191 | 1 | ▲ | ▲ | ▲ a | |||||||||||
| Sum | 191 | 4 | 17 | 29 | 40 | 5 | 38 | 2 | 32 | 93 | 103 | 57 | 3 | 3 | |
| No. | Heart | Liver | Spleen | Lung | Kindey | Brain | Stomach | Intestine |
|---|---|---|---|---|---|---|---|---|
| TAX | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ |
| M2 | - | ▲ | - | ▲ | ▲ | ▲ | ▲ | ▲ |
| M5 | - | ▲ | - | - | ▲ | - | ▲ | - |
| M7 | - | - | - | - | - | - | - | ▲ |
| M11 | ▲ | ▲ | ▲ | ▲ | ▲ | - | ▲ | ▲ |
| M18 | ▲ | ▲ | ▲ | ▲ | ▲ | - | ▲ | ▲ |
| M19 | ▲ | ▲ | ▲ | ▲ | ▲ | - | ▲ | ▲ |
| M20 | - | - | ▲ | ▲ | ▲ | - | ▲ | ▲ |
| M21 | - | ▲ | ▲ | ▲ | ▲ | - | ▲ | ▲ |
| M22 | ▲ | - | - | - | - | - | - | - |
| M23 | - | ▲ | - | - | ▲ | - | ▲ | ▲ |
| M24 | - | ▲ | - | - | - | - | - | ▲ |
| M25 | - | ▲ | - | - | ▲ | - | ▲ | ▲ |
| M28 | - | - | - | - | ▲ | - | ▲ | - |
| M29 | - | - | - | - | - | - | - | ▲ |
| M30 | - | - | - | - | ▲ | - | ▲ | ▲ |
| M31 | - | - | - | - | - | - | ▲ | ▲ |
| M32 | - | ▲ | - | - | ▲ | - | - | ▲ |
| M33 | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ |
| M34 | - | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ |
| M35 | - | ▲ | - | - | ▲ | - | ▲ | ▲ |
| M36 | - | - | - | - | - | - | ▲ | - |
| M42 | - | ▲ | - | - | ▲ | - | ▲ | - |
| M43 | ▲ | - | ▲ | ▲ | - | - | - | ▲ |
| M44 | - | ▲ | - | - | ▲ | - | ▲ | - |
| M45 | - | ▲ | - | - | ▲ | - | ▲ | ▲ |
| M48 | - | ▲ | - | ▲ | ▲ | - | ▲ | ▲ |
| M49 | ▲ | ▲ | ▲ | ▲ | ▲ | - | ▲ | ▲ |
| M50 | - | ▲ | ▲ | ▲ | ▲ | - | ▲ | ▲ |
| M51 | - | - | - | - | - | - | - | ▲ |
| M52 | - | ▲ | - | - | ▲ | - | ▲ | ▲ |
| M65 | - | - | - | - | - | - | ▲ | ▲ |
| M70 | - | - | - | - | - | - | ▲ | ▲ |
| M72 | - | - | - | - | ▲ | - | - | ▲ |
| M75 | - | - | - | - | ▲ | - | - | ▲ |
| M80 | - | - | - | - | ▲ | - | - | ▲ |
| M84 | - | ▲ | - | - | ▲ | - | ▲ | ▲ |
| M86 | - | - | - | - | ▲ | - | - | - |
| M87 | - | - | - | - | - | - | - | ▲ |
| M91 | - | - | - | - | - | - | - | ▲ |
| M105 | - | ▲ | - | - | ▲ | - | ▲ | ▲ |
| M109 | - | - | - | - | ▲ | - | - | ▲ |
| M118 | - | - | - | - | - | - | - | ▲ |
| M150 | - | - | - | - | ▲ | - | - | - |
| M151 | - | - | - | - | ▲ | - | - | - |
| M161 | - | - | - | - | - | - | ▲ | - |
| M162 | - | - | - | - | - | - | ▲ | - |
| SUM | 7 | 22 | 10 | 12 | 31 | 3 | 29 | 35 |
| Fragment No. | Count of Metabolites | The Structures of Metabolites | Bioactive Metabolites and Related Pharmacological Effects |
|---|---|---|---|
 | 4 |  | M105 (one metabolite) Antioxidant, Anti-inflammatory, Antitumor, Antimicrobial, Xanthine oxidase inhibitor |
 | 6 |  | M70, M73, M74, M80 (four metabolites) Antioxidant, Anti-inflammatory, Antitumor, Cardioprotective, Antidiabetic, Antimicrobial, Antiviral, Hepatoprotective, Prevention of Alzheimer disease, Immunoregulatory, Xanthine oxidase inhibitor, Neuroprotective |
 | 18 |  | M70, M73, M74, M75, M80, M83, M104, M105 (eight metabolites) Antioxidant, Anti-inflammatory, Antitumor, Cardioprotective, Antidiabetic, Antimicrobial, Antiviral, Hepatoprotective, Prevention of Alzheimer disease, Immunoregulatory, Xanthine oxidase inhibitor, Neuroprotective |
 | 14 |  | M75, M80, M104, M113, M131, M136 (six metabolites) Antioxidant, Anti-inflammatory, Antitumor, Cardioprotective, Antidiabetic, Antimicrobial, Antiviral, Hepatoprotective, Prevention of Alzheimer disease, Immunoregulatory, Xanthine oxidase inhibitor, Neuroprotective |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, P.; Xu, F.; Li, H.-F.; Wang, Y.; Li, F.-C.; Shang, M.-Y.; Liu, G.-X.; Wang, X.; Cai, S.-Q. Detection of 191 Taxifolin Metabolites and Their Distribution in Rats Using HPLC-ESI-IT-TOF-MSn. Molecules 2016, 21, 1209. https://doi.org/10.3390/molecules21091209
Yang P, Xu F, Li H-F, Wang Y, Li F-C, Shang M-Y, Liu G-X, Wang X, Cai S-Q. Detection of 191 Taxifolin Metabolites and Their Distribution in Rats Using HPLC-ESI-IT-TOF-MSn. Molecules. 2016; 21(9):1209. https://doi.org/10.3390/molecules21091209
Chicago/Turabian StyleYang, Ping, Feng Xu, Hong-Fu Li, Yi Wang, Feng-Chun Li, Ming-Ying Shang, Guang-Xue Liu, Xuan Wang, and Shao-Qing Cai. 2016. "Detection of 191 Taxifolin Metabolites and Their Distribution in Rats Using HPLC-ESI-IT-TOF-MSn" Molecules 21, no. 9: 1209. https://doi.org/10.3390/molecules21091209
APA StyleYang, P., Xu, F., Li, H.-F., Wang, Y., Li, F.-C., Shang, M.-Y., Liu, G.-X., Wang, X., & Cai, S.-Q. (2016). Detection of 191 Taxifolin Metabolites and Their Distribution in Rats Using HPLC-ESI-IT-TOF-MSn. Molecules, 21(9), 1209. https://doi.org/10.3390/molecules21091209