Computational Evaluation of Nucleotide Insertion Opposite Expanded and Widened DNA by the Translesion Synthesis Polymerase Dpo4
Abstract
:1. Introduction
2. Results
2.1. Catalytic Palm Domain Does Not Significantly Rearrange for the Replication of (x,y)DNA Bases, with the Exception of Modified Cytosine
2.2. Reaction Parameters for the Catalytic Step Are Unaffected for the Replication of All (x,y)DNA Bases
2.3. d(x,y)N:dNTP Hydrogen Bonding Does Not Significantly Deviate from Canonical Watson-Crick Interactions, with the Exception of dyA and dyC
2.4. Base-Pair, Base-Step and Helical Parameters for (x,y)DNA Deviate from Canonical Values, with Widened Bases Generally Resulting in Greater Deviations than Expanded Bases
2.5. Canonical dN:dNTP Base-Pair Interactions Are Maintained for (x,y)DNA Bases, with the Exception of dyC, dxC and dyA
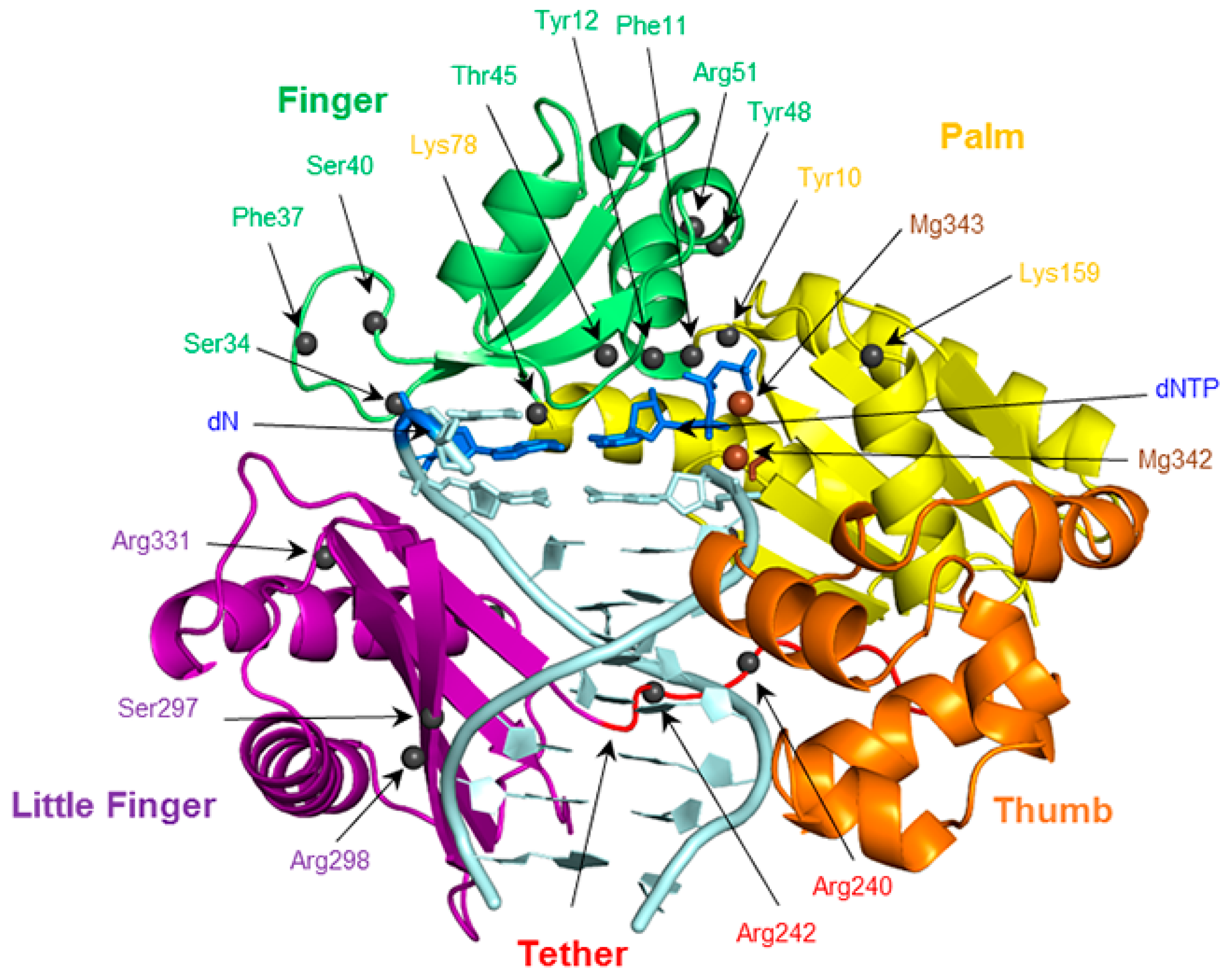
2.6. Residues Interacting with the Template Backbone Move to Accommodate the (x,y)DNA Bases
2.7. Residues in the Ceiling Region of the Finger Domain Rearrange to Accommodate the (x,y)DNA Bases
2.8. Accomodation of the Modified Bases Disrupts the Enzyme Backbone, Particularly for the Finger and Little Finger Domains
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
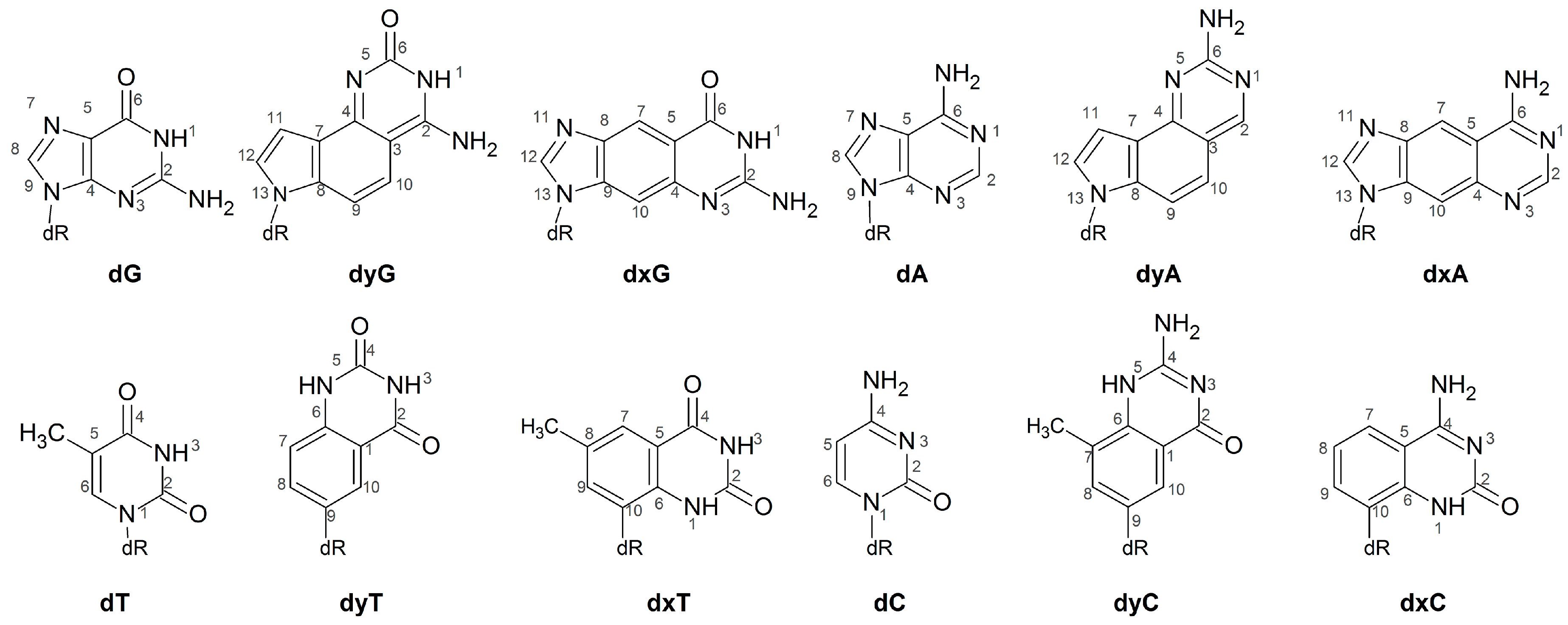
Abbreviations
| Ala | alanine |
| Arg | arginine |
| Asn | asparagine |
| dA | deoxyadenosine monophosphate |
| dC | deoxycytidine monophosphate |
| DNA | deoxyribose nucleic acid |
| dNTP | deoxyribose nucleoside triphosphate |
| Dpo4 | DNA polymerase IV |
| dG | deoxyguanosine monophosphate |
| dN | DNA nucleotide |
| dT | deoxythymidine monophosphate |
| dxA | expanded deoxyadenosine monophosphate |
| dxC | expanded deoxycytidine monophosphate |
| dxG | expanded deoxyguanosine monophosphate |
| dxT | expanded deoxythymidine monophosphate |
| dyA | widened deoxyadenosine monophosphate |
| dyC | widened deoxycytidine monophosphate |
| dyG | widened deoxyguanosine monophosphate |
| dyT | widened deoxythymidine monophosphate |
| εdG | 1,N2-ethenoguanine |
| Glu | glutamine |
| Gly | glycine |
| Kf(exo–) | Klenow Fragment(exo–) |
| Lys | lysine |
| M1dG | malondialdehyde-guanine |
| MD | molecular dynamics |
| Met | methionine |
| Mg | magnesium |
| MM/GBSA | Molecular Mechanics / Generalized Born Surface Area |
| PdG | 1,N2-propanoguanine |
| Phe | phenylalanine |
| RMSD | root-mean-square deviation |
| Ser | serine |
| Thr | threonine |
| Tyr | tyrosine |
| Val | valine |
| xDNA | expanded deoxyribose nucleic acid |
| yDNA | widened deoxyribose nucleic acid |
References
- Webb, S.J. Bioinspired organic chemistry. Annu. Rep. Prog. Chem. Sect. B Org. Chem. 2006, 102, 377–400. [Google Scholar] [CrossRef]
- Cobb, A.J.A. Recent highlights in modified oligonucleotide chemistry. Org. Biomol. Chem. 2007, 5, 3260–3275. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, E.; Peyman, A.; Breipohl, G.; Will, D.W. PNA: Synthetic polyamide nucleic acids with unusual binding properties. Angew. Chem. Int. Ed. Engl. 1998, 37, 2796–2823. [Google Scholar] [CrossRef]
- Kaur, H.; Babu, B.R.; Maiti, S. Perspectives on chemistry and therapeutic applications of locked nucleic acid (LNA). Chem. Rev. 2007, 107, 4672–4697. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, V.B.; Holliger, P. The XNA world: Progress towards replication and evolution of synthetic genetic polymers. Curr. Opin. Chem. Biol. 2012, 16, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Moran, S.; Ren, R.X.F.; Rumney, S.; Kool, E.T. Difluorotoluene, a nonpolar isostere for thymine, codes specifically and efficiently for adenine in DNA replication. J. Am. Chem. Soc. 1997, 119, 2056–2057. [Google Scholar] [CrossRef] [PubMed]
- Egli, M. The steric hypothesis for DNA replication and fluorine hydrogen bonding revisited in light of structural data. Acc. Chem. Res. 2012, 45, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Malyshev, D.A.; Dhami, K.; Quach, H.T.; Lavergne, T.; Ordoukhanian, P.; Torkamani, A.; Romesberg, F.E. Efficient and sequence-independent replication of DNA containing a third base pair establishes a functional six-letter genetic alphabet. Proc. Natl. Acad. Sci. USA 2012, 109, 12005–12010. [Google Scholar] [CrossRef] [PubMed]
- LaPointe, S.M.; Wetmore, S.D. Characterization of the binding interactions between a unique class of modified purines and the natural nucleobases. Chem. Phys. Lett. 2005, 408, 322–328. [Google Scholar] [CrossRef]
- Polak, M.; Seley, K.L.; Plavec, J. Conformational properties of shape modified nucleosides—fleximers. J. Am. Chem. Soc. 2004, 126, 8159–8166. [Google Scholar] [CrossRef] [PubMed]
- Seley, K.L.; Salim, S.; Zhang, L.; O’Daniel, P.I. “Molecular chameleons” design and synthesis of a second series of flexible nucleosides. J. Org. Chem. 2005, 70, 1612–1619. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I.; Kimoto, M.; Yamashige, R. Natural versus artificial creation of base pairs in DNA: Origin of nucleobases from the perspectives of unnatural base pair studies. Acc. Chem. Res. 2012, 45, 2055–2065. [Google Scholar] [CrossRef] [PubMed]
- Miguel, F.-C.; Vincent, M.; Bobby, G.S. Benzo-homologated nucleobases in a nanotube-electrode set-up for DNA sequencing. Nanotechnology 2007, 18, 424019. [Google Scholar]
- Sun, H.; Ren, J.; Qu, X. Carbon nanomaterials and DNA: From molecular recognition to applications. Acc. Chem. Res. 2016, 49, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Deleavey, G.F.; Damha, M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012, 19, 937–954. [Google Scholar] [CrossRef] [PubMed]
- Sahu, N.K.; Shilakari, G.; Nayak, A.; Kohli, D.V. Antisense technology: A selective tool for gene expression regulation and gene targeting. Curr. Pharm. Biotechnol. 2007, 8, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Linko, V.; Ora, A.; Kostiainen, M.A. DNA nanostructures as smart drug-delivery vehicles and molecular devices. Trends Biotechnol. 2015, 33, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Hean, J.; Crowther, C.; Ely, A.; Ul Islam, R.; Barichievy, S.; Bloom, K.; Weinberg, M.S.; van Otterlo, W.A.; de Koning, C.B.; Salazar, F.; et al. Inhibition of hepatitis B virus replication in vivo using lipoplexes containing altritol-modified antiviral sirnas. Artif. DNA PNA XNA 2010, 1, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Marimani, M.; Hean, J.; Bloom, K.; Ely, A.; Arbuthnot, P. Recent advances in developing nucleic acid-based HBV therapy. Future Microbiol. 2013, 8, 1489–1504. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Degardin, M.; Lavergne, T.; Malyshev, D.A.; Dhami, K.; Ordoukhanian, P.; Romesberg, F.E. Natural-like replication of an unnatural base pair for the expansion of the genetic alphabet and biotechnology applications. J. Am. Chem. Soc. 2014, 136, 826–829. [Google Scholar] [CrossRef] [PubMed]
- Teller, C.; Willner, I. Functional nucleic acid nanostructures and DNA machines. Curr. Opin. Biotechnol. 2010, 21, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M. Xenobiology: A new form of life as the ultimate biosafety tool. Bioessays 2010, 32, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, F.; Alvarado, J.B.; Benner, S.A. Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J. Am. Chem. Soc. 2011, 133, 15105–15112. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmsson, L.M. Fluorescent nucleic acid base analogues. Q. Rev. Biophys. 2010, 43, 159–183. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, T.M.; Leonard, N.J. Defined dimensional alterations in enzyme substrates. Lin-naphthoadenine and lin-naphthoadenosine. J. Org. Chem. 1984, 49, 2158–2164. [Google Scholar] [CrossRef]
- Leonard, N.J. Dimensional probes of enzyme-cobinding sites. Acc. Chem. Res. 1982, 15, 128–135. [Google Scholar] [CrossRef]
- Leonard, N.J.; Scopes, D.I.C.; Van der Lijn, P.; Barrio, J.R. Dimensional probes of the enzyme binding sites of adenine nucleotides. Biological effects of widening the adenine ring by 2.4 Å. Biochemistry 1978, 17, 3677–3685. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; He, K.; Kool, E.T. yDNA: A new geometry for size-expanded base pairs. Angew. Chem. Int. Ed. Engl. 2004, 43, 5834–5836. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.; Kool, E.T. Novel benzopyrimidines as widened analogues of DNA bases. J. Org. Chem. 2005, 70, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Lynch, S.R.; Lee, A.H.; Kool, E.T. Structure and replication of yDNA: A novel genetic set widened by benzo-homologation. ChemBioChem 2009, 10, 2530–2538. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.F.; Kool, E.T. A new four-base genetic helix, yDNA, composed of widened benzopyrimidine-purine pairs. J. Am. Chem. Soc. 2005, 127, 3332–3338. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, J.; Kool, E.T. Size-expanded analogues of dG and dC: Synthesis and pairing properties in DNA. J. Org. Chem. 2005, 70, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Krueger, A.T.; Lu, H.; Lee, A.H.F.; Kool, E.T. Synthesis and properties of size-expanded DNAs: Toward designed, functional genetic systems. Acc. Chem. Res. 2007, 40, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lynch, S.R.; Kool, E.T. Solution structure of xDNA: A paired genetic helix with increased diameter. J. Am. Chem. Soc. 2004, 126, 6900–6905. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, J.; Kool, E.T. Helix-forming properties of size-expanded DNA, an alternative four-base genetic form. J. Am. Chem. Soc. 2005, 127, 1396–1402. [Google Scholar] [CrossRef] [PubMed]
- Krueger, A.T.; Kool, E.T. Fluorescence of size-expanded DNA bases: Reporting on DNA sequence and structure with an unnatural genetic set. J. Am. Chem. Soc. 2008, 130, 3989–3899. [Google Scholar] [CrossRef] [PubMed]
- Winnacker, M.; Kool, E.T. Artificial genetic sets composed of size-expanded base pairs. Angew. Chem. Int. Ed. Engl. 2013, 52, 12498–12508. [Google Scholar] [CrossRef] [PubMed]
- McConnell, T.L.; Wetmore, S.D. How do size-expanded DNA nucleobases enhance duplex stability? Computational analysis of the hydrogen-bonding and stacking ability of xDNA bases. J. Phys. Chem. B 2007, 111, 2999–3009. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Cabrera, M.; Zhao, X.; Kent, P.R.C.; Sumpter, B.G. Electronic structure of xDNA. J. Phys. Chem. B 2007, 111, 9057–9061. [Google Scholar] [CrossRef] [PubMed]
- Migliore, A.; Corni, S.; Varsano, D.; Klein, M.L.; Di, F.R. First principles effective electronic couplings for hole transfer in natural and size-expanded DNA. J. Phys. Chem. B 2009, 113, 9402–9415. [Google Scholar] [CrossRef] [PubMed]
- Blas, J.R.; Huertas, O.; Tabares, C.; Sumpter, B.G.; Fuentes-Cabrera, M.; Orozco, M.; Ordejon, P.; Luque, F.J. Structural, dynamical, and electronic transport properties of modified DNA duplexes containing size-expanded nucleobases. J. Phys. Chem. A 2011, 115, 11344–11354. [Google Scholar] [CrossRef] [PubMed]
- Jarchow-Choy, S.K.; Krueger, A.T.; Liu, H.; Gao, J.; Kool, E.T. Fluorescent xDNA nucleotides as efficient substrates for a template-independent polymerase. Nucleic Acids Res. 2011, 39, 1586–1594. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Cabrera, M.; Sumpter, B.G.; Lipkowski, P.; Wells, J.C. Size-expanded yDNA bases: An ab initio study. J. Phys. Chem. B 2006, 110, 6379–6384. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Cabrera, M.; Sumpter, B.G.; Wells, J.C. Size-expanded DNA bases: An ab initio study of their structural and electronic properties. J. Phys. Chem. B 2005, 109, 21135–21139. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Cabrera, M.; Lipkowski, P.; Huertas, O.; Sumpter, B.G.; Orozco, M.; Luque, F.J.; Wells, J.C.; Leszczynski, J. Aromaticity-induced changes in the electronic properties of size-expanded DNA bases: Case of xC. Int. J. Quantum Chem. 2006, 106, 2339–2346. [Google Scholar] [CrossRef]
- Sundaralingam, M.; Ponnuswamy, P.K. Stability of DNA duplexes with Watson-Crick base pairs: A predicted model. Biochemistry 2004, 43, 16467–16476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, H.; Chen, X.; Cukier, R.I.; Bu, Y. Absorption and fluorescence emission spectroscopic characters of size-expanded yDNA bases and effect of deoxyribose and base pairing. J. Phys. Chem. B 2009, 113, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Krueger, A.T. Part I: Fluorescence of Size-Expanded DNA Bases: Reporting on DNA Sequence and Structure with an Unnatural Genetic Set. Part II: Toward Replication of xDNA, a Size-Expanded, Unnatural Genetic System; Stanford University: Stanford, CA, USA, 2009. [Google Scholar]
- Gao, J.; Liu, H.; Kool, E.T. Assembly of the complete eight-base artificial genetic helix, xDNA, and its interaction with the natural genetic system. Angew. Chem. Int. Ed. Engl. 2005, 44, 3118–3122. [Google Scholar] [CrossRef] [PubMed]
- Sale, J.E.; Lehmann, A.R.; Woodgate, R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012, 13, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Voet, D.; Voet, J.G. Biochemistry, 4th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Fijalkowska, I.J.; Schaaper, R.M.; Jonczyk, P. DNA replication fidelity in Escherichia coli: A multi-DNA polymerase affair. FEMS Microbiol. Rev. 2012, 36, 1105–1121. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, S.; Dunn, M.R.; Chaput, J.C. An efficient and faithful in vitro replication system for threose nucleic acid. J. Am. Chem. Soc. 2013, 135, 3583–3591. [Google Scholar] [CrossRef] [PubMed]
- Loakes, D.; Holliger, P. Polymerase engineering: Towards the encoded synthesis of unnatural biopolymers. Chem. Commun. 2009, 4619–4631. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Hongdilokkul, N.; Liu, Z.; Adhikary, R.; Tsuen, S.S.; Romesberg, F.E. Evolution of thermophilic DNA polymerases for the recognition and amplification of C2’-modified DNA. Nat. Chem. 2016, 8, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Chelliserrykattil, J.; Lu, H.; Lee, A.H.; Kool, E.T. Polymerase amplification, cloning, and gene expression of benzo-homologous “yDNA” base pairs. Chembiochem. 2008, 9, 2976–2980. [Google Scholar] [CrossRef] [PubMed]
- Krueger, A.T.; Peterson, L.W.; Chelliserry, J.; Kleinbaum, D.J.; Kool, E.T. Encoding phenotype in bacteria with an alternative genetic set. J. Am. Chem. Soc. 2011, 133, 18447–18451. [Google Scholar] [CrossRef] [PubMed]
- Delaney, J.C.; Gao, J.; Liu, H.; Shrivastav, N.; Essigmann, J.M.; Kool, E.T. Efficient replication bypass of size-expanded DNA base pairs in bacterial cells. Angew. Chem. Int. Ed. Engl. 2009, 48, 4524–4527. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Krueger, A.T.; Gao, J.; Liu, H.; Kool, E.T. Toward a designed genetic system with biochemical function: Polymerase synthesis of single and multiple size-expanded DNA base pairs. Org. Biomol. Chem. 2010, 8, 2704–2710. [Google Scholar] [CrossRef] [PubMed]
- Fiala, K.A.; Suo, Z. Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry 2004, 43, 2106–2115. [Google Scholar] [CrossRef] [PubMed]
- Mizukami, S.; Kim, T.W.; Helquist, S.A.; Kool, E.T. Varying DNA base-pair size in subangstrom increments: Evidence for a loose, not large, active site in low-fidelity Dpo4 polymerase. Biochemistry 2006, 45, 2772–2778. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.C.; Kool, E.T. Varied molecular interactions at the active sites of several DNA polymerases: Nonpolar nucleoside isosteres as probes. J. Am. Chem. Soc. 2000, 122, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Oum, L.; Lee, Y.C.; Geacintov, N.E.; Broyde, S. Visualizing sequence-governed nucleotide selectivities and mutagenic consequences through a replicative cycle: Processing of a bulky carcinogen N2-dG lesion in a Y-family DNA polymerase. Biochemistry 2009, 48, 4677–4690. [Google Scholar] [CrossRef] [PubMed]
- Rechkoblit, O.; Malinina, L.; Cheng, Y.; Kuryavyi, V.; Broyde, S.; Geacintov, N.E.; Patel, D.J. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxog lesion. PLoS Biol. 2006, 4, e11. [Google Scholar] [CrossRef] [PubMed]
- Perlow-Poehnelt, R.A.; Likhterov, I.; Scicchitano, D.A.; Geacintov, N.E.; Broyde, S. The spacious active site of a Y-family DNA polymerase facilitates promiscuous nucleotide incorporation opposite a bulky carcinogen-DNA adduct: Elucidating the structure-function relationship through experimental and computational approaches. J. Biol. Chem. 2004, 279, 36951–336961. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Oum, L.; Geacintov, N.E.; Broyde, S. Nucleotide selectivity opposite a benzo[a]pyrene-derived N2-dG adduct in a Y-family DNA polymerase: A 5′-slippage mechanism. Biochemistry 2008, 47, 2701–2709. [Google Scholar] [CrossRef] [PubMed]
- Brenlla, A.; Markiewicz, R.P.; Rueda, D.; Romano, L.J. Nucleotide selection by the Y-family DNA polymerase Dpo4 involves template translocation and misalignment. Nucleic Acids Res. 2014, 42, 2555–2563. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.; Banerjee, S.; Johnson Salyard, T.L.; Malik, C.K.; Christov, P.P.; Rizzo, C.J.; Stone, M.P.; Egli, M. Structural basis for error-free bypass of the 5-N-methylformamidopyrimidine-dG lesion by human DNA polymerase η and Sulfolobus solfataricus P2 polymerase IV. J. Am. Chem. Soc. 2015, 137, 7011–7014. [Google Scholar] [CrossRef] [PubMed]
- Perlow-Poehnelt, R.A.; Likhterov, I.; Wang, L.; Scicchitano, D.A.; Geacintov, N.E.; Broyde, S. Increased flexibility enhances misincorporation: Temperature effects on nucleotide incorporation opposite a bulky carcinogen-DNA adduct by a Y-family DNA polymerase. J. Biol. Chem. 2007, 282, 1397–1408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Rechkoblit, O.; Wang, L.; Patel, D.J.; Shapiro, R.; Broyde, S. Mutagenic nucleotide incorporation and hindered translocation by a food carcinogen C8-dG adduct in Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): Modeling and dynamics studies. Nucleic Acids Res. 2006, 34, 3326–3337. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Broyde, S. A new anti conformation for N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (AAF-dG) allows Watson-Crick pairing in the Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4). Nucleic Acids Res. 2006, 34, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Chandani, S.; Loechler, E.L. Molecular modeling benzo[a]pyrene N2-dG adducts in the two overlapping active sites of the Y-family DNA polymerase Dpo4. J. Mol. Graph. Model. 2007, 25, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Maxwell, B.A.; Brown, J.A.; Zhang, L.; Suo, Z. Global conformational dynamics of a Y-family DNA polymerase during catalysis. PLoS Biol. 2009, 7, e1000225. [Google Scholar] [CrossRef] [PubMed]
- Nair, D.T.; Kottur, J.; Sharma, R. A rescue act: Translesion DNA synthesis past N2-deoxyguanosine adducts. IUBMB Life 2015, 67, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, M.; Yan, S.F.; Patel, D.J.; Geacintov, N.E.; Broyde, S. Accommodation of a 1S-(−)-benzo[c]phenanthrenyl-N6-dA adduct in the Y-family Dpo4 DNA polymerase active site: Structural insights through molecular dynamics simulations. Chem. Res. Toxicol. 2005, 18, 441–456. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, X.; Hu, P.; Broyde, S.; Zhang, Y. A water-mediated and substrate-assisted catalytic mechanism for Sulfolobus solfataricus DNA polymerase IV. J. Am. Chem. Soc. 2007, 129, 4731–4737. [Google Scholar] [CrossRef] [PubMed]
- Vaisman, A.; Ling, H.; Woodgate, R.; Yang, W. Fidelity of Dpo4: Effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005, 24, 2957–2967. [Google Scholar] [CrossRef] [PubMed]
- Gaur, V.; Vyas, R.; Fowler, J.D.; Efthimiopoulos, G.; Feng, J.Y.; Suo, Z. Structural and kinetic insights into binding and incorporation of l-nucleotide analogs by a Y-family DNA polymerase. Nucleic Acids Res. 2014, 42, 9984–9995. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.M.; Parasuram, R.; Rajput, P.R.; Rozners, E.; Ondrechen, M.J.; Beuning, P.J. Effects of non-catalytic, distal amino acid residues on activity of E. Coli DinB (DNA polymerase IV). Environ. Mol. Mutagen. 2012, 53, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Jarosz, D.F.; Godoy, V.G.; Delaney, J.C.; Essigmann, J.M.; Walker, G.C. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 2006, 439, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Arora, K.; Schlick, T. Subtle but variable conformational rearrangements in the replication cycle of Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) may accommodate lesion bypass. Protein Sci. 2006, 15, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Schlick, T. Quantum mechanics/molecular mechanics investigation of the chemical reaction in Dpo4 reveals water-dependent pathways and requirements for active site reorganization. J. Am. Chem. Soc. 2008, 130, 13240–13250. [Google Scholar] [CrossRef] [PubMed]
- Washington, M.T.; Helquist, S.A.; Kool, E.T.; Prakash, L.; Prakash, S. Requirement of Watson-Crick hydrogen bonding for DNA synthesis by yeast DNA polymerase η. Mol. Cell. Biol. 2003, 23, 5107–5112. [Google Scholar] [CrossRef] [PubMed]
- Wolfle, W.T.; Washington, M.T.; Kool, E.T.; Spratt, T.E.; Helquist, S.A.; Prakash, L.; Prakash, S. Evidence for a Watson-Crick hydrogen bonding requirement in DNA synthesis by human DNA polymerase κ. Mol. Cell. Biol. 2005, 25, 7137–7143. [Google Scholar] [CrossRef] [PubMed]
- Kool, E.T. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 2002, 71, 191–219. [Google Scholar] [CrossRef] [PubMed]
- Kool, E.T.; Morales, J.C.; Guckian, K.M. Mimicking the structure and function of DNA: Insights into DNA stability and replication. Angew. Chem. Int. Ed. Engl. 2000, 39, 990–1009. [Google Scholar] [CrossRef]
- Liljas, A.; Liljas, L.; Piškur, J.; Lindblom, G.; Nissen, P.; Kjeldgaard, M. Textbook of Structural Biology; World Scientific: Singapore, Singapore, 2009. [Google Scholar]
- Lu, X.-J.; Olson, W.K. 3DNA: A versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008, 3, 1213–1227. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.; Jayaram, B. Energetics of base pairs in B-DNA in solution: An appraisal of potential functions and dielectric treatments. J. Phys. Chem. B 1998, 102, 6139–6144. [Google Scholar] [CrossRef]
- Sponer, J.; Jurecka, P.; Marchan, I.; Luque, F.J.; Orozco, M.; Hobza, P. Nature of base stacking: Reference quantum-chemical stacking energies in ten unique B-DNA base-pair steps. Chem. Eur. J. 2006, 12, 2854–2865. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Boudsocq, F.; Woodgate, R.; Yang, W. Crystal structure of a Y-family DNA polymerase in action: A mechanism for error-prone and lesion-bypass replication. Cell 2001, 107, 91–102. [Google Scholar] [CrossRef]
- Battersby, T.R.; Albalos, M.; Friesenhahn, M.J. An unusual mode of DNA duplex association: Watson-Crick interaction of all-purine deoxyribonucleic acids. Chem. Biol. 2007, 14, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Buckley, R.; Enekwa, C.D.; Williams, L.D.; Hud, N.V. Molecular recognition of watson-crick-like purine-purine base pairs. ChemBioChem 2011, 12, 2155–2158. [Google Scholar] [CrossRef] [PubMed]
- Heuberger, B.D.; Switzer, C. An alternative nucleobase code: Characterization of purine-purine DNA double helices bearing guanine-isoguanine and diaminopurine 7-deaza-xanthine base pairs. Chembiochem. 2008, 9, 2779–2783. [Google Scholar] [CrossRef] [PubMed]
- Cafferty, B.J.; Hud, N.V. Was a pyrimidine-pyrimidine base pair the ancestor of Watson-Crick base pairs? Insights from a systematic approach to the origin of RNA. Isr. J. Chem. 2015, 55, 891–905. [Google Scholar] [CrossRef]
- Wächtershäuser, G. An all-purine precursor of nucleic acids. Proc. Natl. Acad. Sci. USA 1988, 85, 1134–1135. [Google Scholar] [CrossRef] [PubMed]
- Zang, H.; Goodenough, A.K.; Choi, J.-Y.; Irimia, A.; Loukachevitch, L.V.; Kozekov, I.D.; Angel, K.C.; Rizzo, C.J.; Egli, M.; Guengerich, F.P. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: Analysis and crystal structures of multiple base pair subsitutions and frameshift products with the adducts 1,N2-ethenoguanine. J. Biol. Chem. 2005, 280, 29750–29764. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Musser, S.K.; Saleh, S.; Marnett, L.J.; Egli, M.; Stone, M.P. Insertion of dntps opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry 2008, 47, 7322–7334. [Google Scholar] [CrossRef] [PubMed]
- Eoff, R.L.; Stafford, J.B.; Szekely, J.; Rizzo, C.J.; Egli, M.; Guengerich, F.P.; Marnett, L.J. Structural and functional analysis of Sulfolobus solfataricus Y-family DNA polymerase Dpo4-catalyzed bypass of the malondialdehyde-deoxyguanosine adduct. Biochemistry 2009, 48, 7079–7088. [Google Scholar] [CrossRef] [PubMed]
- Gahlon, H.L.; Schweizer, W.B.; Sturla, S.J. Tolerance of base pair size and shape in postlesion DNA synthesis. J. Am. Chem. Soc. 2013, 135, 6384–6387. [Google Scholar] [CrossRef] [PubMed]
- Gahlon, H.L.; Boby, M.L.; Sturla, S.J. O6-alkylguanine postlesion DNA synthesis is correct with the right complement of hydrogen bonding. ACS Chem. Biol. 2014, 9, 2807–2814. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Sayer, J.M.; Plosky, B.S.; Yagi, H.; Boudsocq, F.; Woodgate, R.; Jerina, D.M.; Yang, W. Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc. Natl. Acad. Sci. USA 2004, 101, 2265–2269. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; T.E. Cheatham, I.; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. Amber 14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Merz, K.M. Taking into account the ion-induced dipole interaction in the nonbonded model of ions. J. Chem. Theory Comput. 2014, 10, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general Amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Perlow, R.A.; Broyde, S. Toward understanding the mutagenicity of an environmental carcinogen: Structural insights into nucleotide incorporation preferences. J. Mol. Biol. 2002, 322, 291–309. [Google Scholar] [CrossRef]
- Zhang, L.; Shapiro, R.; Broyde, S. Molecular dynamics of a food carcinogen-DNA adduct in a replicative DNA polymerase suggest hindered nucleotide incorporation and extension. Chem. Res. Toxicol. 2005, 18, 1347–1363. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Revision D.01; Gaussian, Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Dupradeau, F.-Y.; Pigache, A.; Zaffran, T.; Savineau, C.; Lelong, R.; Grivel, N.; Lelong, D.; Rosanski, W.; Cieplak, P. The R.E.D. Tools: Advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010, 12, 7821–7839. [Google Scholar] [CrossRef] [PubMed]
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.-Y. R.E.D. Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011, 39, W511–W517. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Becker, J.-P.; Cieplak, P.; Dupradeau, F.-Y. Red Python: Object Oriented Programming for Amber Force Fields; American Chemical Society: Washington, DC, USA, 2014. [Google Scholar]
- Bayly, C.I.; Cieplak, P.; Cornell, W.; Kollman, P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Lavery, R.; Moakher, M.; Maddocks, J.H.; Petkeviciute, D.; Zakrzewska, K. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 2009, 37, 5917–5929. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are not available from the authors.
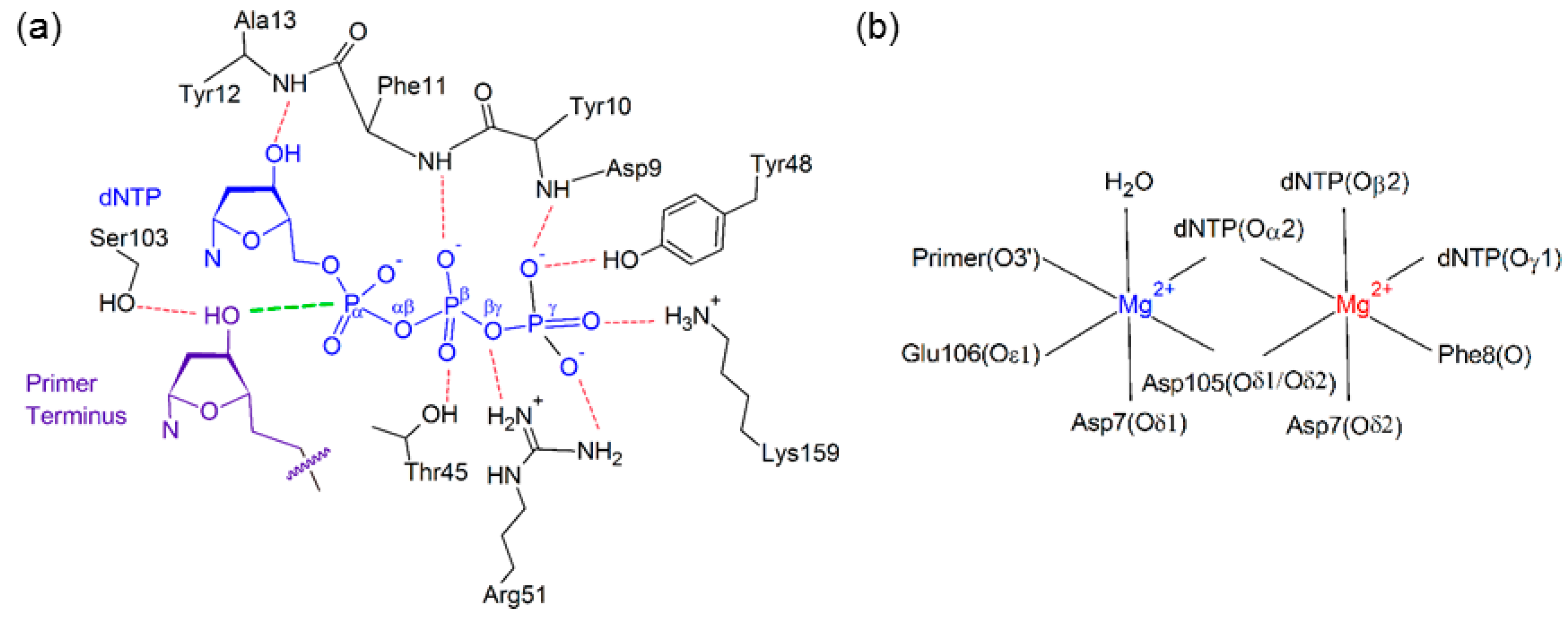
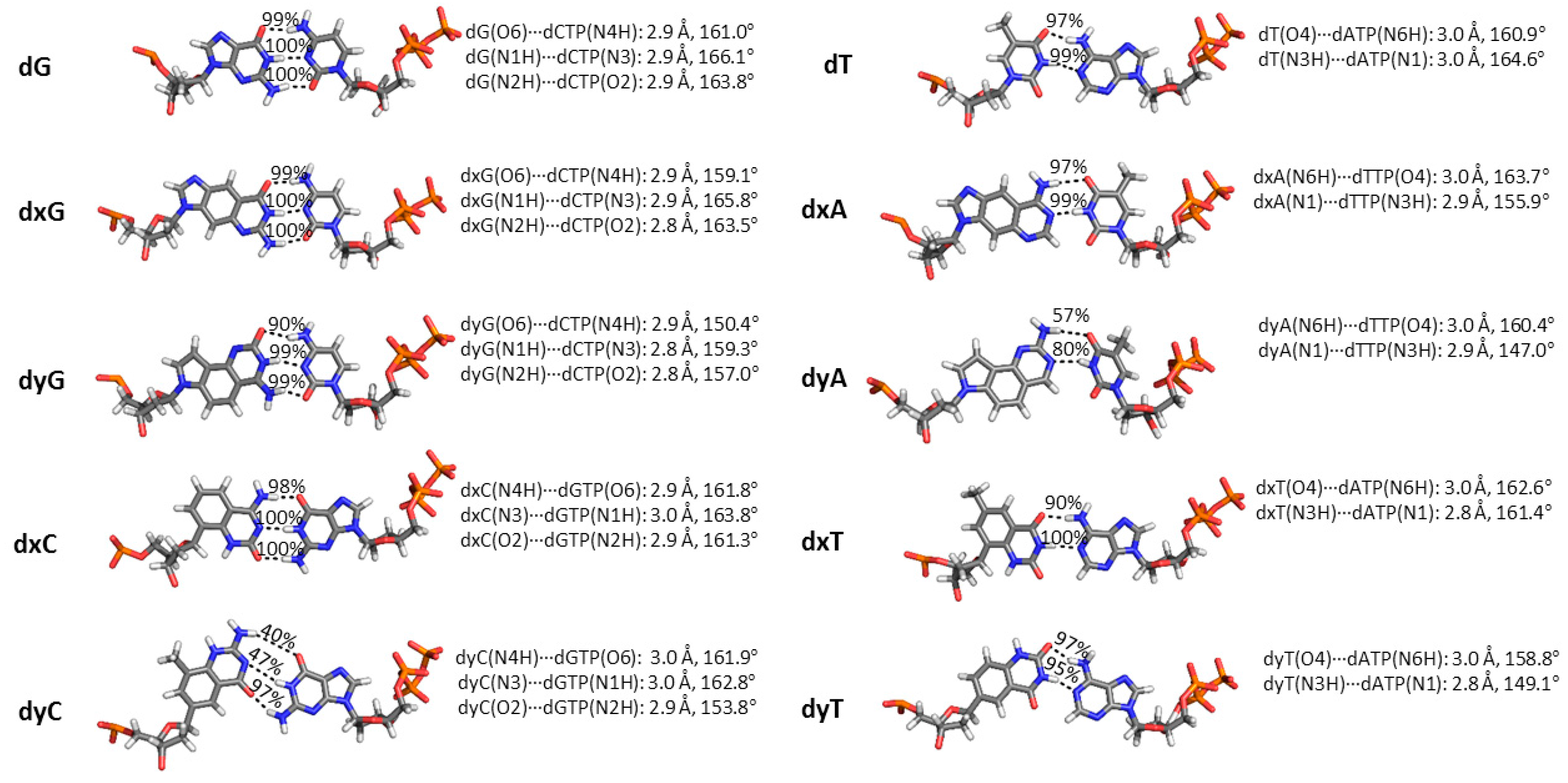




| Active Site Base Pair | dNTP(Oβγ) Arg51(Nη1H) | dNTP(Oγ) Arg51(Nη2H) | dNTP(Oγ) Lys159(NζH) | dNTP(Oγ) Tyr10(NH) | dNTP(Oβ) Thr45(OγH) | dNTP(Oβ) dNTP(O3′H) |
|---|---|---|---|---|---|---|
| dG:dCTP | 100% | 97% | 90% | 98% | 99% | 8% |
| dT:dATP | 98% | 99% | 88% | 99% | 96% | 99% |
| dxA:dTTP | 98% | 100% | 93% | 96% | 98% | 11% |
| dyA:dTTP | 93% | 100% | 76% | 98% | 100% | NA b |
| dxG:dCTP | 100% | 90% | 68% | 98% | 97% | 6% |
| dyG:dCTP | 99% | 90% | 71% | 98% | 95% | NA b |
| dxT:dATP | 99% | 97% | 61% | 99% | 96% | 100% |
| dyT:dATP | 99% | 98% | 78% | 99% | 96% | 100% |
| dxC:dGTP | NA b | 100% | 99% | 44% | NA b | 93% |
| dyC:dGTP | NA b | 100% | 99% | 41% | NA b | 93% |
| Active Site Base Pair | d(O3′–Pα) | ∠(O3′–Pα–Oαβ) |
|---|---|---|
| dG:dCTP | 3.6 ± 0.1 Å | 172.9 ± 3.7° |
| dT:dATP | 3.5 ± 0.1 Å | 174.0 ± 3.0° |
| dxA:dTTP | 3.6 ± 0.1 Å | 174.4 ± 3.0° |
| dyA:dTTP | 3.6 ± 0.2 Å | 174.2 ± 3.1° |
| dxG:dCTP | 3.6 ± 0.1 Å | 174.1 ± 3.2° |
| dyG:dCTP | 3.6 ± 0.1 Å | 174.2 ± 3.1° |
| dxT:dATP | 3.5 ± 0.1 Å | 174.1 ± 3.1° |
| dyT:dATP | 3.5 ± 0.1 Å | 173.7 ± 3.3° |
| dxC:dGTP | 3.6 ± 0.2 Å | 163.1 ± 4.5° |
| dyC:dGTP | 3.6 ± 0.2 Å | 162.6 ± 4.5° |
| Active Site Base Pair | Buckle | Propeller | Opening | Shear | Stretch | C1′–C1′ Distance | Stagger |
|---|---|---|---|---|---|---|---|
| dG:dCTP | –5.3 ± 7.4 | –1.2 ± 7.1 | 0.8 ± 2.9 | 0.2 ± 0.3 | 0.0 ± 0.1 | 10.8 ± 0.1 | –0.3 ± 0.4 |
| dT:dATP | 1.5 ± 7.8 | –4.7 ± 7.1 | 0.6 ± 4.7 | –0.1 ± 0.3 | 0.1 ± 0.1 | 10.8 ± 0.2 | –0.1 ± 0.4 |
| dxA:dTTP | –14.2 ± 9.3 | –9.5 ± 7.1 | 5.5 ± 5.0 | 0.4 ± 0.3 | 2.2 ± 0.2 | 12.6 ± 0.2 | –1.2 ± 0.7 |
| dyA:dTTP | –28.7 ± 13.6 | –19.8 ± 9.5 | 3.7 ± 12.2 | –2.6 ± 0.6 | 1.2 ± 0.3 | 11.9 ± 0.5 | –1.9 ± 0.9 |
| dxG:dCTP | –18.2 ± 9.6 | –6.0 ± 7.2 | 1.5 ± 3.5 | 0.4 ± 0.3 | 2.3 ± 0.1 | 12.8 ± 0.2 | –1.3 ± 0.6 |
| dyG:dCTP | –30.6 ± 10.2 | –10.1 ± 7.9 | –3.3 ± 3.5 | –1.3 ± 0.4 | 1.7 ± 0.1 | 12.4 ± 0.2 | –1.7 ± 0.7 |
| dxT:dATP | –8.5 ± 7.8 | –7.2 ± 9.3 | 1.3 ± 5.9 | 0.4 ± 0.3 | 1.8 ± 0.2 | 12.2 ± 0.2 | –0.4 ± 0.5 |
| dyT:dATP | –24.2 ± 13.1 | –2.7 ± 7.5 | 5.1 ± 3.7 | –1.2 ± 0.3 | 1.9 ± 0.2 | 12.2 ± 0.3 | –0.1 ± 0.7 |
| dxC:dGTP | 3.3 ± 6.8 | –6.3 ± 9.1 | 3.1 ± 3.8 | 1.2 ± 0.3 | 2.1 ± 0.1 | 12.7 ± 0.2 | 0.3 ± 0.4 |
| dyC:dGTP | 0.1 ± 9.3 | –15.1 ± 8.5 | 13.8 ± 14.0 | –0.3 ± 0.9 | 2.4 ± 0.4 | 12.4 ± 0.5 | –0.2 ± 0.4 |
| Active Site Base Pair | Tilt | Roll | Twist | Shift | Slide | Rise | Inclination | Tip | Major Groove Width | Minor Groove Width | Axial Bend |
|---|---|---|---|---|---|---|---|---|---|---|---|
| dG:dCTP | –4.1 ± 3.5 | 7.7 ± 4.1 | 26.3 ± 3.1 | 0.2 ± 0.4 | –0.8 ± 0.3 | 3.2 ± 0.2 | 18.2 ± 4.5 | 0.1 ± 4.4 | 14.0 ± 1.4 | 8.3 ± 1.8 | 2.2 ± 1.0 |
| dT:dATP | –0.7 ± 4.0 | 5.0 ± 4.7 | 29.8 ± 3.1 | 0.4 ± 0.5 | –0.9 ± 0.3 | 3.2 ± 0.3 | 14.0 ± 5.7 | 0.1 ± 4.2 | 13.4 ± 1.6 | 8.0 ± 2.0 | 2.0 ± 0.9 |
| dxA:dTTP | –7.7 ± 4.2 | 4.9 ± 3.8 | 21.9 ± 2.8 | 0.1 ± 0.5 | –1.2 ± 0.3 | 3.1 ± 0.2 | 17.8 ± 5.7 | 4.6 ± 4.5 | 13.2 ± 0.9 | 7.8 ± 1.7 | 2.2 ± 1.1 |
| dyA:dTTP | –17.3 ± 7.5 | 4.8 ± 4.6 | 36.1 ± 4.0 | 0.0 ± 0.7 | 0.0 ± 0.8 | 2.9 ± 0.3 | 23.3 ± 6.5 | 12.4 ± 5.5 | 13.5 ± 1.0 | 6.4 ± 3.7 | 3.7 ± 2.0 |
| dxG:dCTP | –8.0 ± 4.3 | 6.2 ± 4.1 | 22.1 ± 3.0 | –0.1 ± 0.4 | –1.2 ± 0.3 | 3.1 ± 0.2 | 20.5 ± 5.3 | 3.9 ± 4.6 | 13.3 ± 1.0 | 7.6 ± 2.7 | 3.2 ± 1.5 |
| dyG:dCTP | –12.4 ± 5.2 | 4.6 ± 3.7 | 28.2 ± 3.0 | –0.1 ± 0.4 | –0.6 ± 0.4 | 3.0 ± 0.3 | 20.2 ± 4.8 | 8.6 ± 4.6 | 13.2 ± 1.1 | 8.5 ± 1.9 | 2.1 ± 1.1 |
| dxT:dATP | –1.8 ± 4.0 | 5.8 ± 4.4 | 25.1 ± 3.2 | 0.3 ± 0.5 | –1.4 ± 0.3 | 3.1 ± 0.2 | 15.0 ± 5.4 | –1.1 ± 4.3 | 13.5 ± 1.0 | 8.1 ± 1.3 | 2.7 ± 1.2 |
| dyT:dATP | –4.4 ± 4.8 | 1.4 ± 5.2 | 29.9 ± 3.0 | –0.2 ± 0.4 | –0.9 ± 0.4 | 3.3 ± 0.3 | 12.0 ± 5.3 | 4.1 ± 5.0 | 13.3 ± 1.2 | 8.2 ± 1.4 | 2.5 ± 1.2 |
| dxC:dGTP | –5.8 ± 3.3 | 5.7 ± 4.1 | 19.8 ± 3.4 | 0.2 ± 0.4 | –1.5 ± 0.3 | 3.4 ± 0.2 | 19.7 ± 14.6 | –1.9 ± 14.7 | 13.1 ± 0.9 | 7.9 ± 1.1 | 2.6 ± 1.1 |
| dyC:dGTP | –6.8 ± 3.8 | 10.0 ± 5.0 | 26.5 ± 3.5 | –0.6 ± 0.8 | –0.9 ± 0.4 | 3.3 ± 0.2 | 22.0 ± 7.0 | 0.9 ± 4.6 | 13.6 ± 1.2 | 8.4 ± 2.0 | 1.9 ± 1.0 |
| Active Site Base Pair | dN:dNTP Hydrogen Bonding | dC:dG n−1 Hydrogen Bonding | dNTP–dG n−1 Stacking | dN–dC n−1 Stacking | dN–dC n+1 Stacking | dN:dNTP–dC:dG n−1 Stacking |
|---|---|---|---|---|---|---|
| dG:dCTP | −30.7 ± 2.7 | −29.3 ± 2.5 | −6.1 ± 0.6 | −6.6 ± 0.6 | −0.3 ± 0.2 | −15.4 ± 1.0 |
| dT:dATP | −7.0 ± 1.6 | −28.8 ± 2.7 | −7.1 ± 0.6 | −5.1 ± 0.7 | −0.4 ± 0.2 | −15.1 ± 0.9 |
| dxA:dTTP | −9.0 ± 2.0 | −27.9 ± 3.0 | −6.6 ± 0.6 | −7.3 ± 0.7 | −0.4 ± 0.3 | −16.8 ± 1.0 |
| dyA:dTTP | −5.8 ± 3.5 | −27.6 ± 3.0 | −5.5 ± 0.9 | −6.9 ± 0.7 | −0.4 ± 0.4 | −15.6 ± 1.3 |
| dxG:dCTP | −26.9 ± 2.7 | −28.3 ± 2.8 | −6.0 ± 0.6 | −7.6 ± 0.7 | −1.5 ± 1.1 | −16.8 ± 1.0 |
| dyG:dCTP | −31.4 ± 3.8 | −28.0 ± 2.7 | −5.7 ± 0.6 | −6.9 ± 0.7 | −0.5 ± 0.4 | −15.7 ± 1.0 |
| dxT:dATP | −9.4 ± 1.6 | −27.6 ± 3.2 | −6.9 ± 0.7 | −4.6 ± 0.7 | −1.9 ± 1.4 | −14.0 ± 1.1 |
| dyT:dATP | −9.3 ± 1.7 | −27.6 ± 3.0 | −6.9 ± 0.8 | −4.1 ± 0.8 | −0.6 ± 0.7 | −13.4 ± 1.2 |
| dxC:dGTP | −23.4 ± 2.6 | −27.8 ± 3.0 | −7.2 ± 0.7 | −5.5 ± 0.7 | 0.0 ± 0.1 | −15.6 ± 1.1 |
| dyC:dGTP | −4.5 ± 2.9 | −27.9 ± 2.8 | −7.6 ± 0.8 | −5.4 ± 0.9 | −3.3 ± 1.7 | −14.4 ± 1.3 |
| Active Site Base Pair | dN | Template n−1 | dNTP | Primer n−1 | Ser34 | Ser40 | Gly58 | Met76 | Arg331 |
|---|---|---|---|---|---|---|---|---|---|
| dxA:dTTP | 1.7 ± 0.5 | 1.0 ± 0.3 | 0.5 ± 0.1 | 0.7 ± 0.5 | 1.1 ± 0.4 | 1.1 ± 0.4 | 0.8 ± 0.4 | 0.7 ± 0.3 | 1.4 ± 0.4 |
| dyA:dTTP | 1.2 ± 0.4 | 0.7 ± 0.3 | 0.4 ± 0.1 | 0.8 ± 0.5 | 1.1 ± 0.4 | 1.0 ± 0.4 | 0.9 ± 0.4 | 0.7 ± 0.3 | 0.8 ± 0.4 |
| dxG:dCTP | 1.8 ± 0.4 | 1.0 ± 0.3 | 0.5 ± 0.1 | 0.7 ± 0.4 | 1.1 ± 0.4 | 0.9 ± 0.5 | 0.9 ± 0.4 | 0.7 ± 0.3 | 1.1 ± 0.4 |
| dyG:dCTP | 1.4 ± 0.3 | 0.9 ± 0.3 | 0.5 ± 0.2 | 0.8 ± 0.3 | 1.1 ± 0.4 | 1.1 ± 0.4 | 0.8 ± 0.4 | 0.7 ± 0.3 | 1.2 ± 0.4 |
| dxT:dATP | 3.0 ± 0.8 | 1.4 ± 0.4 | 0.4 ± 0.2 | 0.8 ± 0.4 | 0.9 ± 0.4 | 1.0 ± 0.5 | 1.0 ± 0.4 | 0.7 ± 0.3 | 1.3 ± 0.4 |
| dyT:dATP | 2.6 ± 1.0 | 1.0 ± 0.4 | 0.5 ± 0.2 | 0.8 ± 0.4 | 1.1 ± 0.5 | 1.2 ± 0.6 | 1.0 ± 0.4 | 0.8 ± 0.4 | 1.3 ± 0.5 |
| dxC:dGTP | 2.6 ± 0.9 | 0.8 ± 0.4 | 0.4 ± 0.2 | 0.8 ± 0.4 | 1.3 ± 0.4 | 1.1 ± 0.4 | 1.3 ± 0.5 | 0.9 ± 0.4 | 1.1 ± 0.4 |
| dyC:dGTP | 2.5 ± 0.8 | 0.8 ± 0.3 | 0.4 ± 0.1 | 0.8 ± 0.4 | 1.1 ± 0.4 | 1.2 ± 0.5 | 1.2 ± 0.5 | 0.7 ± 0.3 | 1.2 ± 0.4 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albrecht, L.; Wilson, K.A.; Wetmore, S.D. Computational Evaluation of Nucleotide Insertion Opposite Expanded and Widened DNA by the Translesion Synthesis Polymerase Dpo4. Molecules 2016, 21, 822. https://doi.org/10.3390/molecules21070822
Albrecht L, Wilson KA, Wetmore SD. Computational Evaluation of Nucleotide Insertion Opposite Expanded and Widened DNA by the Translesion Synthesis Polymerase Dpo4. Molecules. 2016; 21(7):822. https://doi.org/10.3390/molecules21070822
Chicago/Turabian StyleAlbrecht, Laura, Katie A. Wilson, and Stacey D. Wetmore. 2016. "Computational Evaluation of Nucleotide Insertion Opposite Expanded and Widened DNA by the Translesion Synthesis Polymerase Dpo4" Molecules 21, no. 7: 822. https://doi.org/10.3390/molecules21070822
APA StyleAlbrecht, L., Wilson, K. A., & Wetmore, S. D. (2016). Computational Evaluation of Nucleotide Insertion Opposite Expanded and Widened DNA by the Translesion Synthesis Polymerase Dpo4. Molecules, 21(7), 822. https://doi.org/10.3390/molecules21070822






