Methods for Improving Aptamer Binding Affinity
Abstract
:1. Introduction
2. From Sequence to Function
2.1. Sequence to Structure
2.2. Structure to Function
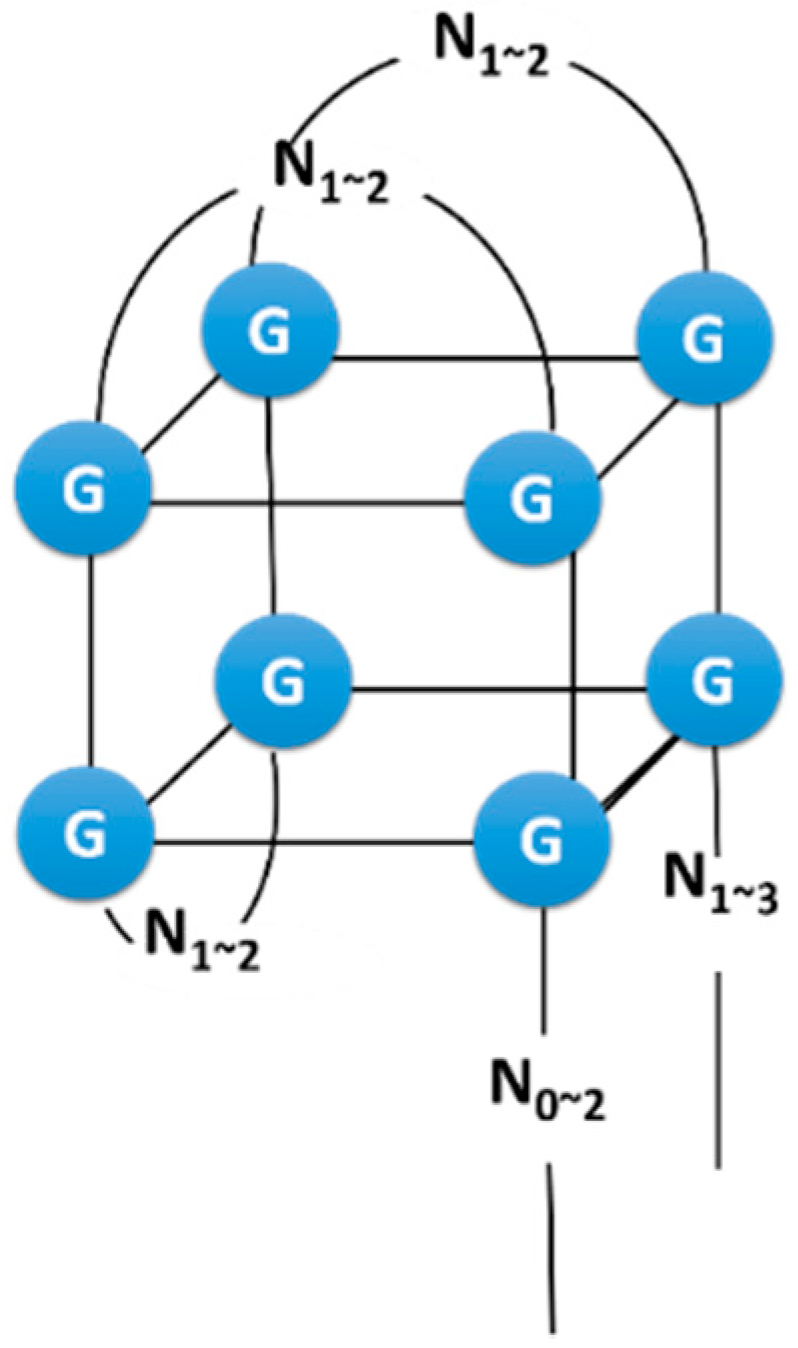
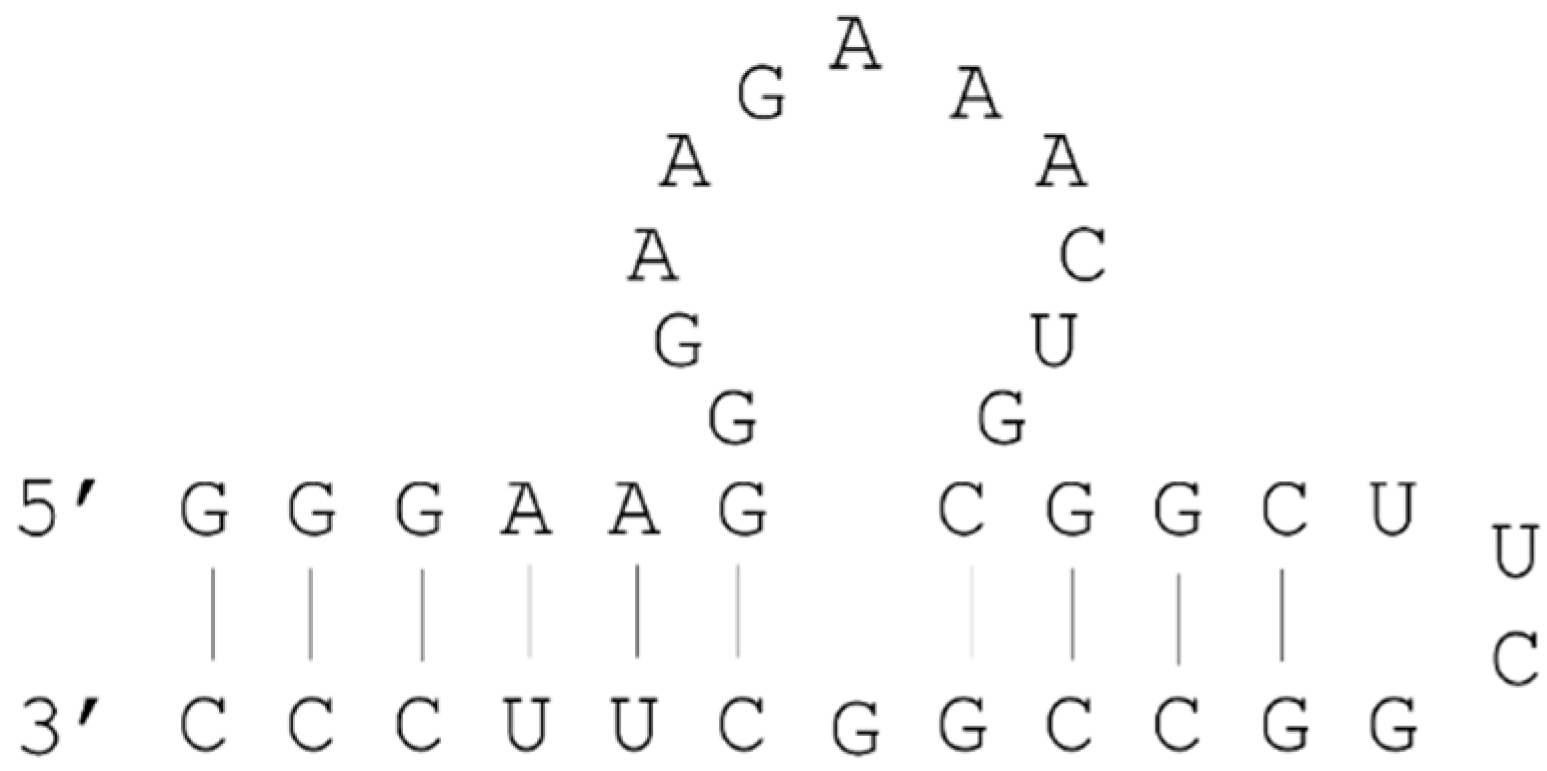
2.3. Example: ATP Aptamer
3. Tuning of Aptamer Binding
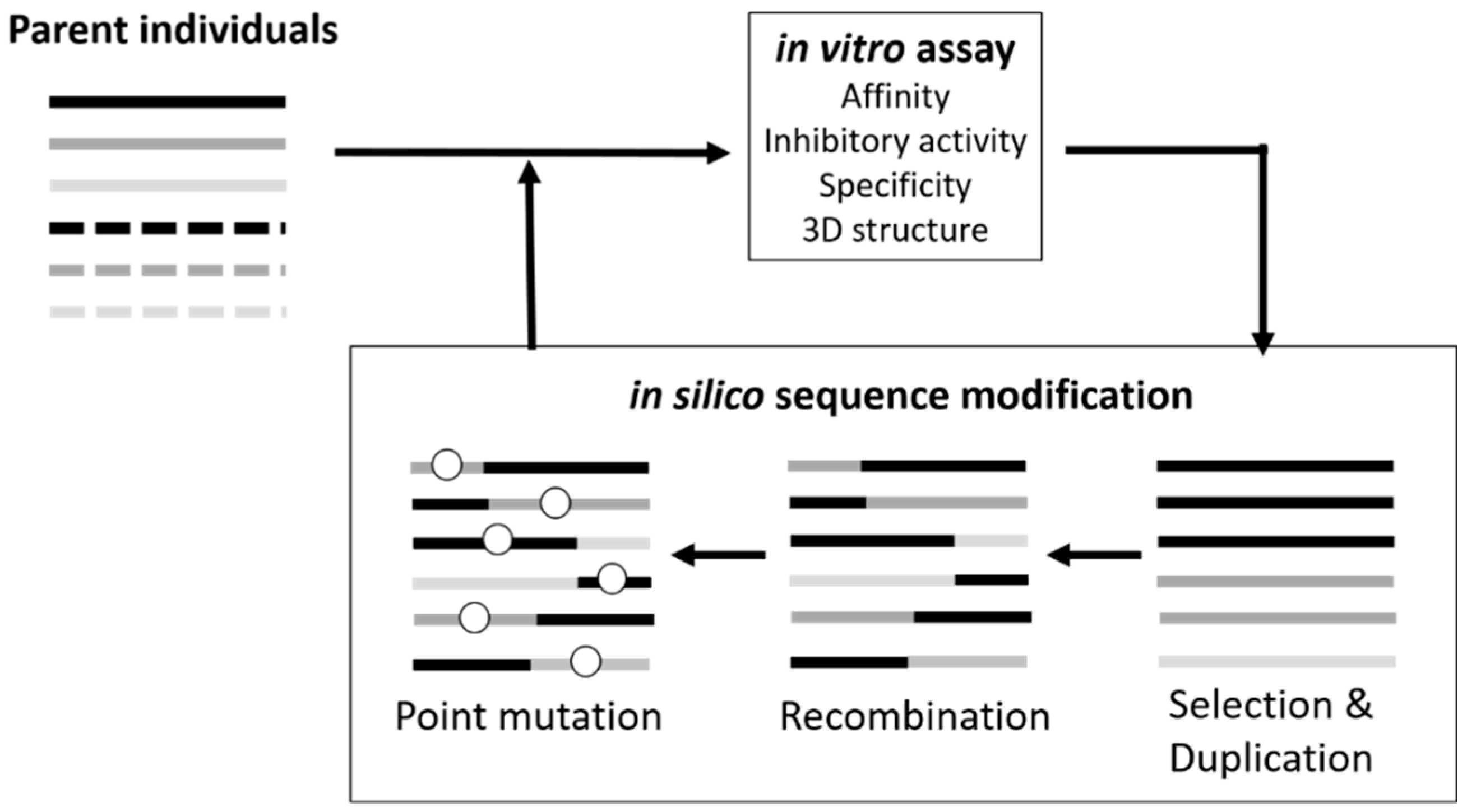
3.1. Sequence Optimization
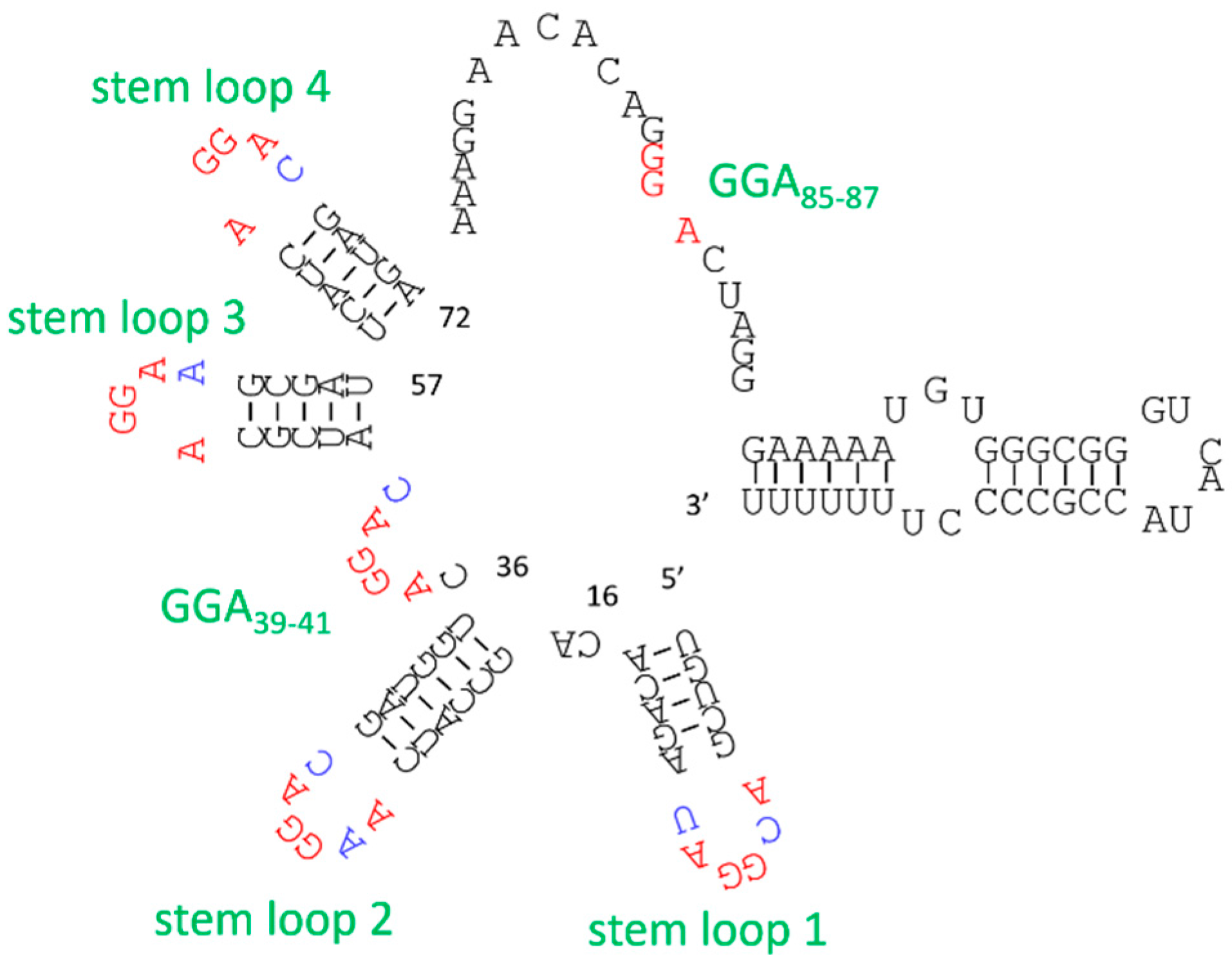
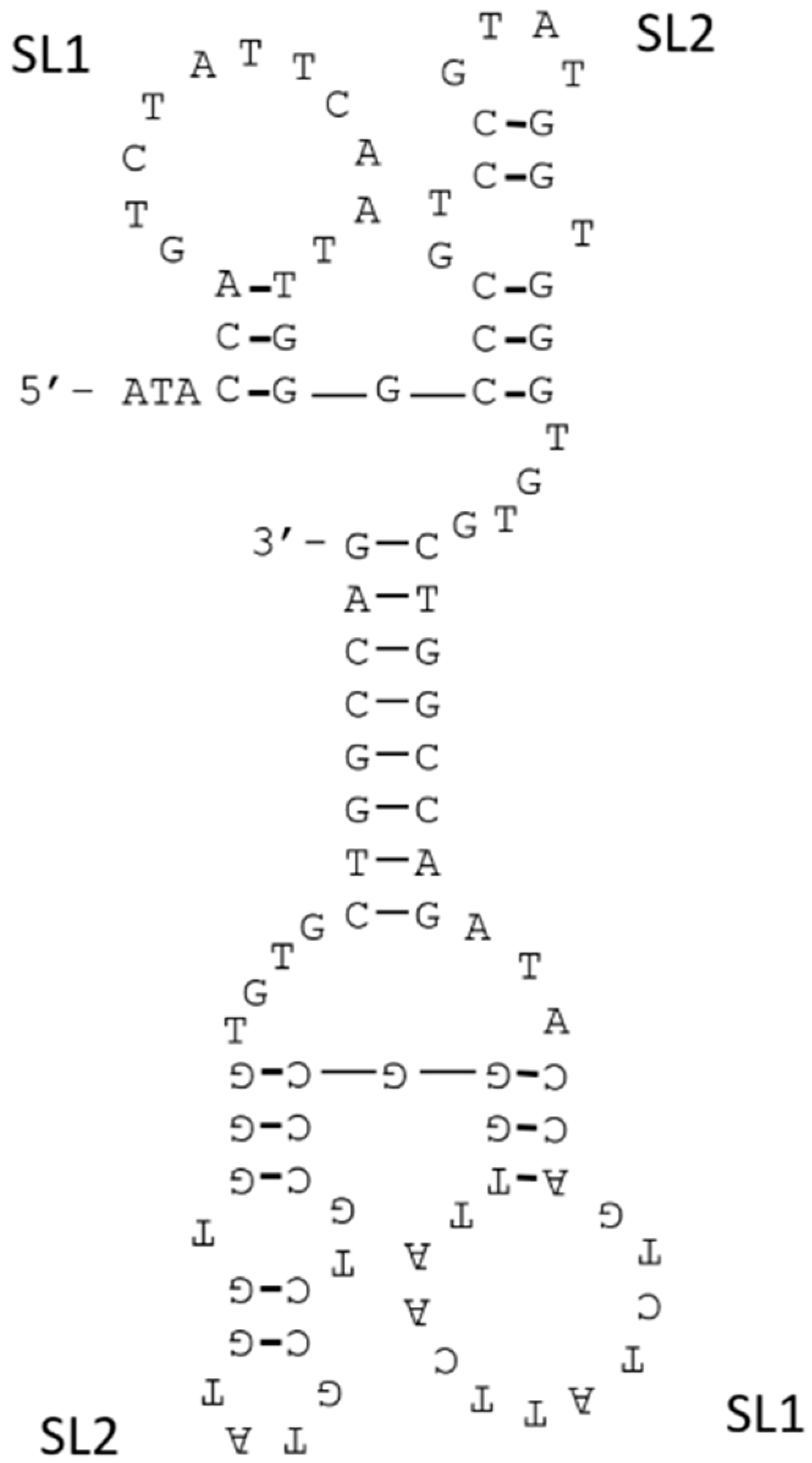
3.2. Structure Manipulation
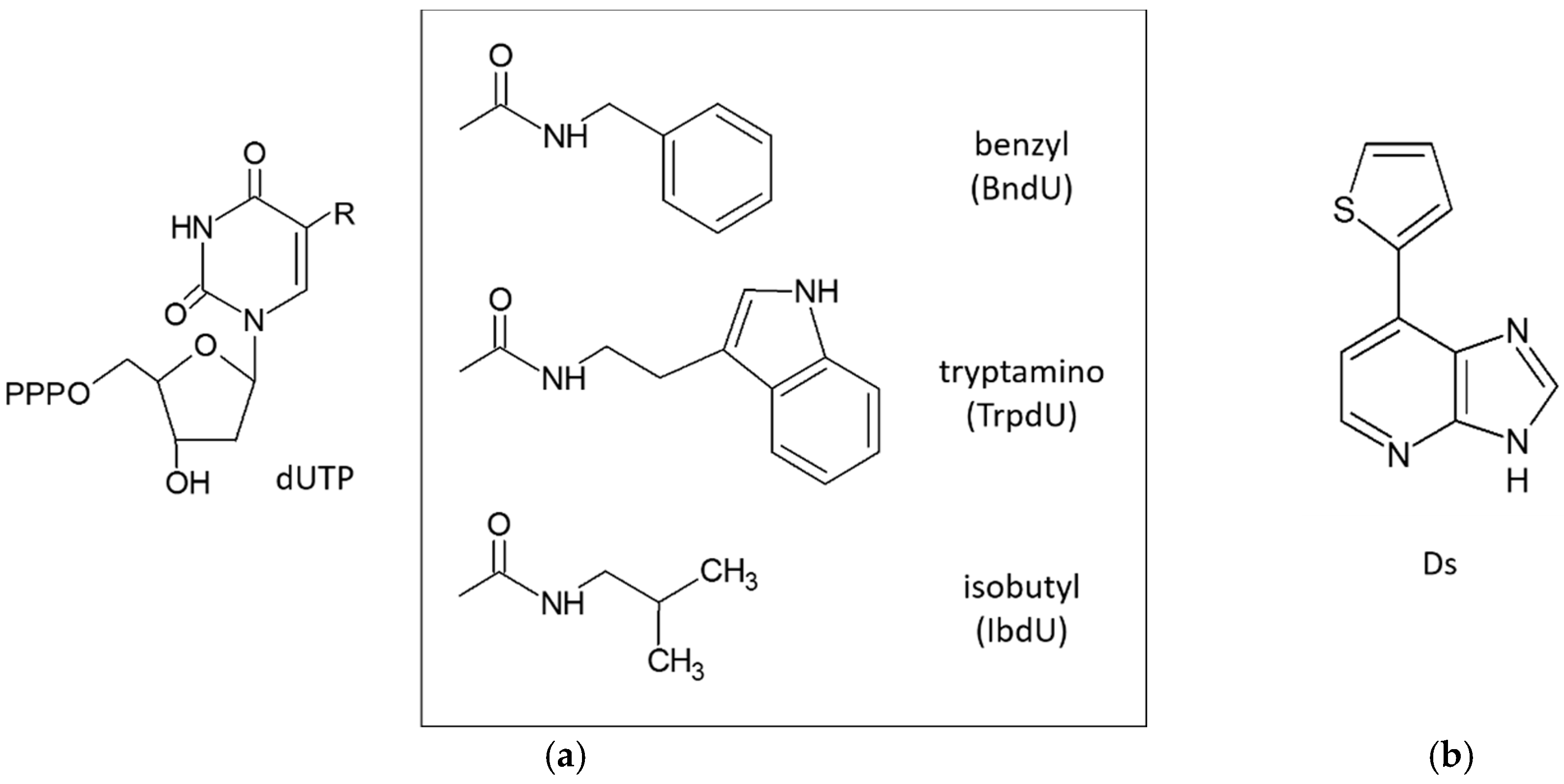
3.3. Extended Alphabet (Artificial Nucleotides)
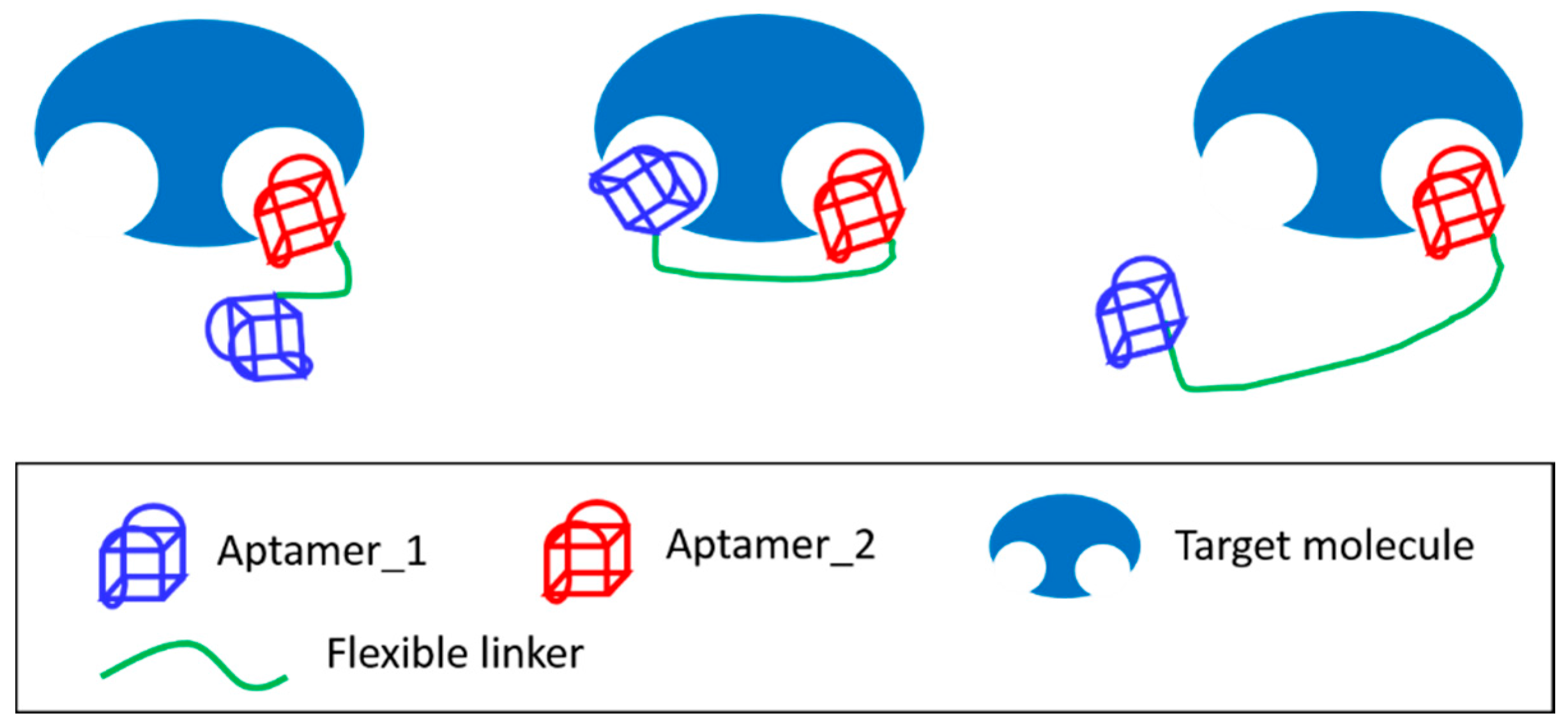
3.4. Joining of Binding Motifs
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of rna molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Hasegawa, H.; Ikebukuro, K. Electrochemical detection of vascular endothelial growth factor by an aptamer-based bound/free separation system. Electrochemistry 2012, 80, 348–352. [Google Scholar] [CrossRef]
- Lee, K.Y.; Kang, H.; Ryu, S.H.; Lee, D.S.; Lee, J.H.; Kim, S. Bioimaging of nucleolin aptamer-containing 5-(n-benzylcarboxyamide)-2′-deoxyuridine more capable of specific binding to targets in cancer cells. J. Biomed. Biotechnol. 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, M.; Yoshida, W.; Yamazaki, H.; Sode, K.; Ikebukuro, K. An aptamer-based bound/free separation system for protein detection. Electroanalysis 2009, 21, 1297–1302. [Google Scholar] [CrossRef]
- Min, K.; Jo, H.; Song, K.; Cho, M.; Chun, Y.S.; Jon, S.; Kim, W.J.; Ban, C. Dual-aptamer-based delivery vehicle of doxorubicin to both PSMA (+) and PSMA (−) prostate cancers. Biomaterials 2011, 32, 2124–2132. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, W.; Sode, K.; Ikebukuro, K. Aptameric enzyme subunit for biosensing based on enzymatic activity measurement. Anal. Chem. 2006, 78, 3296–3303. [Google Scholar] [CrossRef] [PubMed]
- Schutze, T.; Rubelt, F.; Repkow, J.; Greiner, N.; Erdmann, V.A.; Lehrach, H.; Konthur, Z.; Glokler, J. A streamlined protocol for emulsion polymerase chain reaction and subsequent purification. Anal. Biochem. 2011, 410, 155–157. [Google Scholar] [CrossRef] [PubMed]
- Shao, K.; Ding, W.; Wang, F.; Li, H.; Ma, D.; Wang, H. Emulsion PCR: A high efficient way of PCR amplification of random DNA libraries in aptamer selection. PLoS ONE 2011, 6, e24910. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjic, N. 2′-fluoropyrimidine rna-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and vegf-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar] [CrossRef] [PubMed]
- Tasset, D.M.; Kubik, M.F.; Steiner, W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 1997, 272, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Sode, K.; Ikebukuro, K. Selection of DNA aptamers against VEGF165 using a protein competitor and the aptamer blotting method. Biotechnol. Lett. 2008, 30, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Yung, L.Y. Probing high affinity sequences of DNA aptamer against VEGF165. PLoS ONE 2012, 7, e31196. [Google Scholar] [CrossRef] [PubMed]
- Mei, H.; Bing, T.; Yang, X.; Qi, C.; Chang, T.; Liu, X.; Cao, Z.; Shangguan, D. Functional-group specific aptamers indirectly recognizing compounds with alkyl amino group. Anal. Chem. 2012, 84, 7323–7329. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.; Bing, T.; Mei, H.; Yang, X.; Liu, X.; Shangguan, D. G-quadruplex DNA aptamers for zeatin recognizing. Biosens. Bioelectron. 2013, 41, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Katilius, E.; Flores, C.; Woodbury, N.W. Exploring the sequence space of a DNA aptamer using microarrays. Nucleic Acids Res. 2007, 35, 7626–7635. [Google Scholar] [CrossRef] [PubMed]
- Tome, J.M.; Ozer, A.; Pagano, J.M.; Gheba, D.; Schroth, G.P.; Lis, J.T. Comprehensive analysis of RNA-protein interactions by high-throughput sequencing-rna affinity profiling. Nat. Methods 2014, 11, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Bing, T.; Yang, X.; Mei, H.; Cao, Z.; Shangguan, D. Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories. Bioorg. Med. Chem. 2010, 18, 1798–1805. [Google Scholar] [CrossRef] [PubMed]
- Sacca, B.; Lacroix, L.; Mergny, J.L. The effect of chemical modifications on the thermal stability of different g-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005, 33, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- Curtis, E.A.; Liu, D.R. Discovery of widespread gtp-binding motifs in genomic DNA and RNA. Chem. Biol. 2013, 20, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Lauhon, C.T.; Szostak, J.W. Rna aptamers that bind flavin and nicotinamide redox cofactors. J. Am. Chem. Soc. 1995, 117, 1246–1257. [Google Scholar] [CrossRef] [PubMed]
- Dieckmann, T.; Butcher, S.E.; Sassanfar, M.; Szostak, J.W.; Feigon, J. Mutant ATP-binding RNA aptamers reveal the structural basis for ligand binding. J. Mol. Biol. 1997, 273, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Dieckmann, T.; Suzuki, E.; Nakamura, G.K.; Feigon, J. Solution structure of an ATP-binding RNA aptamer reveals a novel fold. RNA 1996, 2, 628–640. [Google Scholar] [PubMed]
- Jiang, F.; Kumar, R.A.; Jones, R.A.; Patel, D.J. Structural basis of RNA folding and recognition in an AMP-RNA aptamer complex. Nature 1996, 382, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Klug, S.J.; Famulok, M. All you wanted to know about selex. Mol. Biol. Rep. 1994, 20, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Crameri, A.; Stemmer, W.P. 10(20)-fold aptamer library amplification without gel purification. Nucleic Acids Res. 1993, 21. [Google Scholar] [CrossRef]
- Kanagawa, T. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 2003, 96, 317–323. [Google Scholar] [CrossRef]
- Yoshida, W.; Mochizuki, E.; Takase, M.; Hasegawa, H.; Morita, Y.; Yamazaki, H.; Sode, K.; Ikebukuro, K. Selection of DNA aptamers against insulin and construction of an aptameric enzyme subunit for insulin sensing. Biosens. Bioelectron. 2009, 24, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.H.; Szostak, J.W. Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl. Acad. Sci. USA 2002, 99, 11616–11621. [Google Scholar] [CrossRef] [PubMed]
- Rowe, W.; Platt, M.; Wedge, D.C.; Day, P.J.; Kell, D.B.; Knowles, J. Analysis of a complete DNA-protein affinity landscape. J. R. Soc. Interface 2010, 7, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Asai, R.; Nishimura, S.I.; Aita, T.; Takahashi, K. In vitro selection of DNA aptamers on chips using a method for generating point mutations. Anal. Lett. 2004, 37, 645–656. [Google Scholar] [CrossRef]
- Ikebukuro, K.; Okumura, Y.; Sumikura, K.; Karube, I. A novel method of screening thrombin-inhibiting DNA aptamers using an evolution-mimicking algorithm. Nucleic Acids Res. 2005, 33, e108. [Google Scholar] [CrossRef] [PubMed]
- Savory, N.; Abe, K.; Yoshida, W.; Ikebukuro, K. In Silico Maturation: Processing Sequences to Improve Biopolymer Functions Based on Genetic Algorithms; Springer International Publishing: Cham, Switzerland, 2014; pp. 273–290. [Google Scholar]
- Holland, J.H. Adaptation in Natural and Artificial Systems: An Introductory Analysis with Applications to Biology, Control, and Artificial Intelligence; University of Michigan Press: Ann Arbor, MI, USA, 1975. [Google Scholar]
- Bittker, J.A.; Le, B.V.; Liu, D.R. Nucleic acid evolution and minimization by nonhomologous random recombination. Nat. Biotechnol. 2002, 20, 1024–1029. [Google Scholar] [CrossRef] [PubMed]
- Ikebukuro, K.; Yoshida, W.; Noma, T.; Sode, K. Analysis of the evolution of the thrombin-inhibiting DNA aptamers using a genetic algorithm. Biotechnol. Lett. 2006, 28, 1933–1937. [Google Scholar] [CrossRef] [PubMed]
- Noma, T.; Ikebukuro, K. Aptamer selection based on inhibitory activity using an evolution-mimicking algorithm. Biochem. Biophys. Res. Commun. 2006, 347, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Fukaya, T.; Abe, K.; Savory, N.; Tsukakoshi, K.; Yoshida, W.; Ferri, S.; Sode, K.; Ikebukuro, K. Improvement of the VEGF binding ability of DNA aptamers through in silico maturation and multimerization strategy. J. Biotechnol. 2015, 212, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Savory, N.; Abe, K.; Sode, K.; Ikebukuro, K. Selection of DNA aptamer against prostate specific antigen using a genetic algorithm and application to sensing. Biosens. Bioelectron. 2010, 26, 1386–1391. [Google Scholar] [CrossRef] [PubMed]
- Savory, N.; Lednor, D.; Tsukakoshi, K.; Abe, K.; Yoshida, W.; Ferri, S.; Jones, B.V.; Ikebukuro, K. In silico maturation of binding-specificity of DNA aptamers against proteus mirabilis. Biotechnol. Bioeng. 2013, 110, 2573–2580. [Google Scholar] [CrossRef] [PubMed]
- Savory, N.; Takahashi, Y.; Tsukakoshi, K.; Hasegawa, H.; Takase, M.; Abe, K.; Yoshida, W.; Ferri, S.; Kumazawa, S.; Sode, K.; et al. Simultaneous improvement of specificity and affinity of aptamers against streptococcus mutans by in silico maturation for biosensor development. Biotechnol. Bioeng. 2014, 111, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Knight, C.G.; Platt, M.; Rowe, W.; Wedge, D.C.; Khan, F.; Day, P.J.; McShea, A.; Knowles, J.; Kell, D.B. Array-based evolution of DNA aptamers allows modelling of an explicit sequence-fitness landscape. Nucleic Acids Res. 2009, 37, e6. [Google Scholar] [CrossRef] [PubMed]
- Platt, M.; Rowe, W.; Wedge, D.C.; Kell, D.B.; Knowles, J.; Day, P.J. Aptamer evolution for array-based diagnostics. Anal. Biochem. 2009, 390, 203–205. [Google Scholar] [CrossRef] [PubMed]
- Rowe, W.; Platt, M.; Wedge, D.C.; Day, P.J.; Kell, D.B.; Knowles, J.D. Convergent evolution to an aptamer observed in small populations on DNA microarrays. Phys. Biol. 2010, 7. [Google Scholar] [CrossRef] [PubMed]
- Koshkin, A.A.; Singh, S.K.; Nielsen, P.; Rajwanshi, V.K.; Kumar, R.; Meldgaard, M.; Olsen, C.E.; Wengel, J. LNA (locked nucleic acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 1998, 54, 3607–3630. [Google Scholar] [CrossRef]
- Schmidt, K.S.; Borkowski, S.; Kurreck, J.; Stephens, A.W.; Bald, R.; Hecht, M.; Friebe, M.; Dinkelborg, L.; Erdmann, V.A. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004, 32, 5757–5765. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, F.J.; Kalra, N.; Wengel, J.; Vester, B. Aptamers as a model for functional evaluation of LNA and 2′-amino LNA. Bioorg. Med. Chem. Lett. 2009, 19, 6585–6587. [Google Scholar] [CrossRef] [PubMed]
- Mallikaratchy, P.R.; Ruggiero, A.; Gardner, J.R.; Kuryavyi, V.; Maguire, W.F.; Heaney, M.L.; McDevitt, M.R.; Patel, D.J.; Scheinberg, D.A. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Res. 2011, 39, 2458–2469. [Google Scholar] [CrossRef] [PubMed]
- Eaton, B.E. The joys of in vitro selection: Chemically dressing oligonucleotides to satiate protein targets. Curr. Opin. Chem. Biol. 1997, 1, 10–16. [Google Scholar] [CrossRef]
- Vaught, J.D.; Bock, C.; Carter, J.; Fitzwater, T.; Otis, M.; Schneider, D.; Rolando, J.; Waugh, S.; Wilcox, S.K.; Eaton, B.E. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 2010, 132, 4141–4151. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Yamashige, R.; Matsunaga, K.; Yokoyama, S.; Hirao, I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013, 31, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.R.; Gelinas, A.D.; Zhang, C.; Rohloff, J.C.; Carter, J.D.; O’Connell, D.; Waugh, S.M.; Wolk, S.K.; Mayfield, W.S.; Burgin, A.B.; et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc. Natl. Acad. Sci. USA 2012, 109, 19971–19976. [Google Scholar] [CrossRef] [PubMed]
- Ochsner, U.A.; Green, L.S.; Gold, L.; Janjic, N. Systematic selection of modified aptamer pairs for diagnostic sandwich assays. Biotechniques 2014, 56, 125–128, 130 and 132–133. [Google Scholar] [PubMed]
- Hasegawa, H.; Taira, K.; Sode, K.; Ikebukuro, K. Improvement of aptamer affinity by dimerization. Sensors 2008, 8, 1090–1098. [Google Scholar] [CrossRef]
- Kim, Y.; Cao, Z.; Tan, W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc. Natl. Acad. Sci. USA 2008, 105, 5664–5669. [Google Scholar] [CrossRef] [PubMed]
- Duss, O.; Michel, E.; Yulikov, M.; Schubert, M.; Jeschke, G.; Allain, F.H. Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature 2014, 509, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Bujotzek, A.; Shan, M.; Haag, R.; Weber, M. Towards a rational spacer design for bivalent inhibition of estrogen receptor. J. Comput. Aided Mol. Des. 2011, 25, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Heyduk, T. Bivalent ligands with long nanometer-scale flexible linkers. Biochemistry 2009, 48, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Kramer, R.H.; Karpen, J.W. Spanning binding sites on allosteric proteins with polymer-linked ligand dimers. Nature 1998, 395, 710–713. [Google Scholar] [CrossRef] [PubMed]
- Fasting, C.; Schalley, C.A.; Weber, M.; Seitz, O.; Hecht, S.; Koksch, B.; Dernedde, J.; Graf, C.; Knapp, E.W.; Haag, R. Multivalency as a chemical organization and action principle. Angew. Chem. Int. Ed. Engl. 2012, 51, 10472–10498. [Google Scholar] [CrossRef] [PubMed]
- Hansma, H.G.; Kim, K.J.; Laney, D.E.; Garcia, R.A.; Argaman, M.; Allen, M.J.; Parsons, S.M. Properties of biomolecules measured from atomic force microscope images: A review. J. Struct. Biol. 1997, 119, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Weers, B.; Stellwagen, N.C. DNA persistence length revisited. Biopolymers 2001, 61, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Lis, J.T.; Shi, H. A systematic study of the features critical for designing a high avidity multivalent aptamer. Nucleic Acid Ther. 2013, 23, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, K.M.; Xiao, Y.; Soh, H.T. Selection is more intelligent than design: Improving the affinity of a bivalent ligand through directed evolution. Nucleic Acids Res. 2012, 40, 11777–11783. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Wang, J.; Ahmad, K.M.; Csordas, A.T.; Zhou, J.; Nie, J.; Stewart, R.; Thomson, J.A.; Rossi, J.J.; Soh, H.T. Selection strategy to generate aptamer pairs that bind to distinct sites on protein targets. Anal. Chem. 2012, 84, 5365–5371. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Lee, S.J.; Choi, E.J.; Kim, J.; Song, J.Y.; Gu, M.B. An ultra-sensitive detection of a whole virus using dual aptamers developed by immobilization-free screening. Biosens. Bioelectron. 2014, 51, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Fan, X.; Sevilimedu, A.; Lis, J.T. RNA aptamers directed to discrete functional sites on a single protein structural domain. Proc. Natl. Acad. Sci. USA 2007, 104, 3742–3746. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, Y.; Sode, K.; Ikebukuro, K. Screening and improvement of an anti-VEGF DNA aptamer. Molecules 2010, 15, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.; Oh, S.S.; Nie, J.; Stewart, R.; Radeke, M.J.; Eisenstein, M.; Coffey, P.J.; Thomson, J.A.; Soh, H.T. Array-based discovery of aptamer pairs. Anal. Chem. 2015, 87, 821–828. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasegawa, H.; Savory, N.; Abe, K.; Ikebukuro, K. Methods for Improving Aptamer Binding Affinity. Molecules 2016, 21, 421. https://doi.org/10.3390/molecules21040421
Hasegawa H, Savory N, Abe K, Ikebukuro K. Methods for Improving Aptamer Binding Affinity. Molecules. 2016; 21(4):421. https://doi.org/10.3390/molecules21040421
Chicago/Turabian StyleHasegawa, Hijiri, Nasa Savory, Koichi Abe, and Kazunori Ikebukuro. 2016. "Methods for Improving Aptamer Binding Affinity" Molecules 21, no. 4: 421. https://doi.org/10.3390/molecules21040421





