Synthesis of N-(6-Arylbenzo[d]thiazole-2-acetamide Derivatives and Their Biological Activities: An Experimental and Computational Approach
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Studies
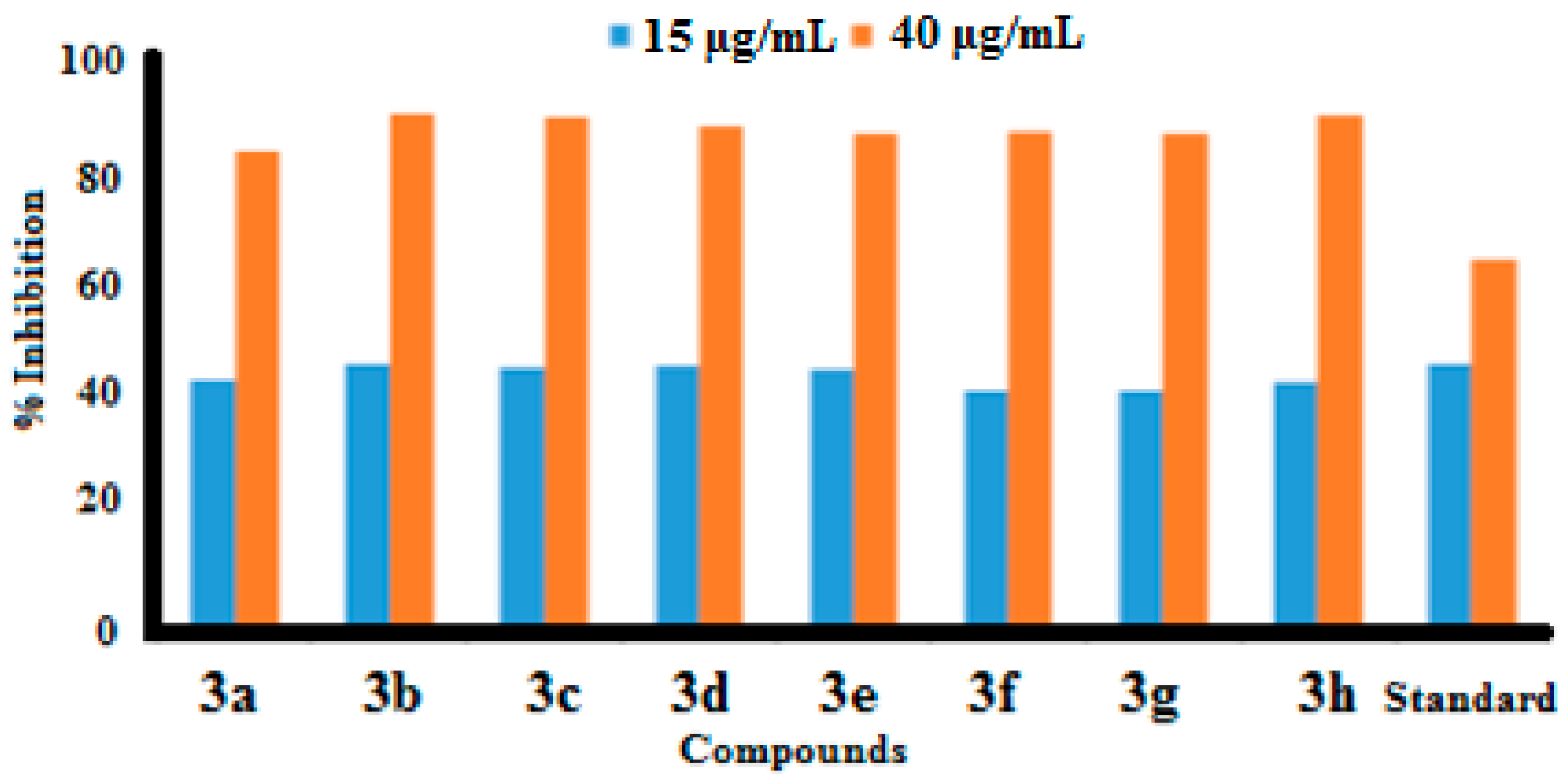
2.2.1. Urease Inhibitory Activity
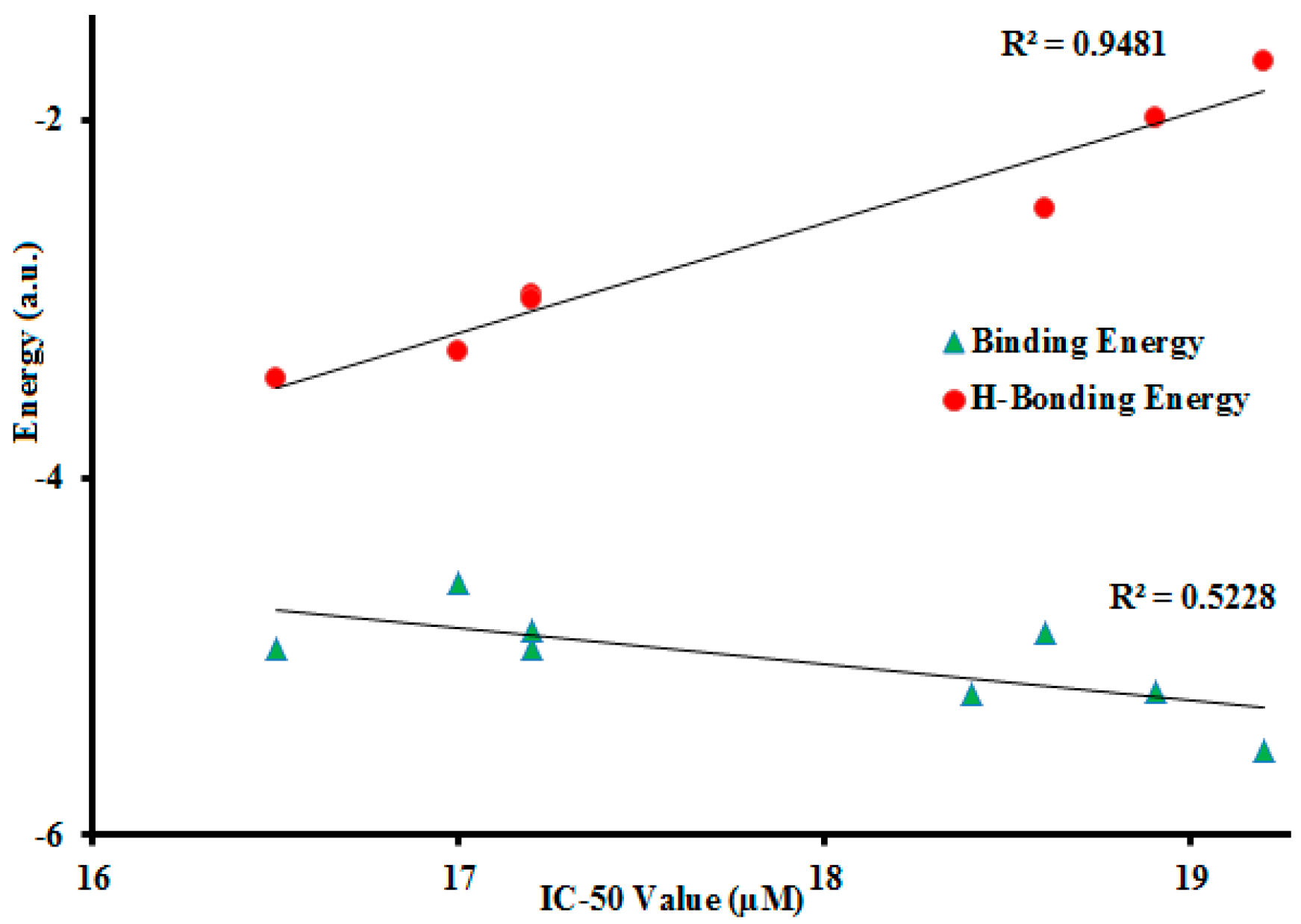
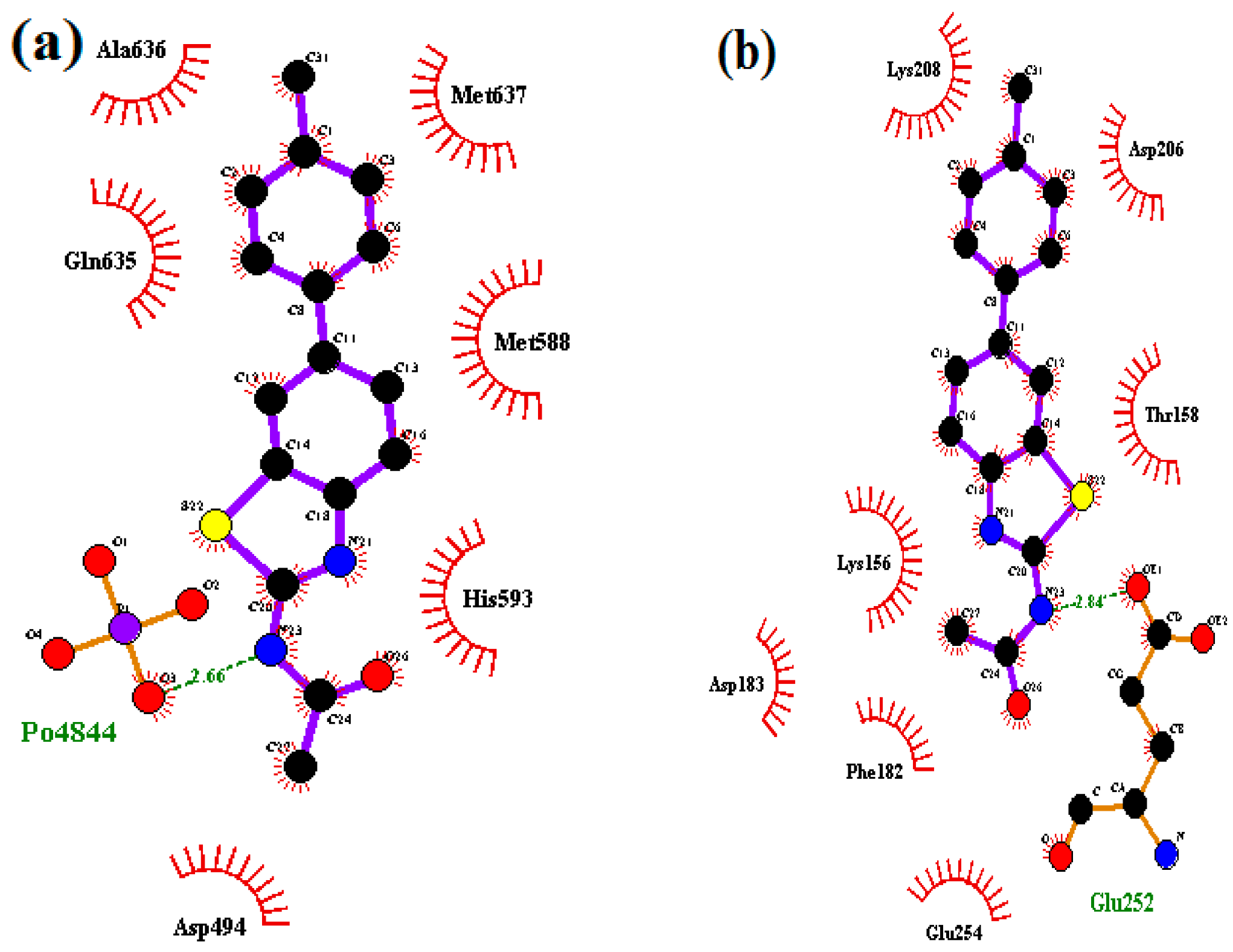
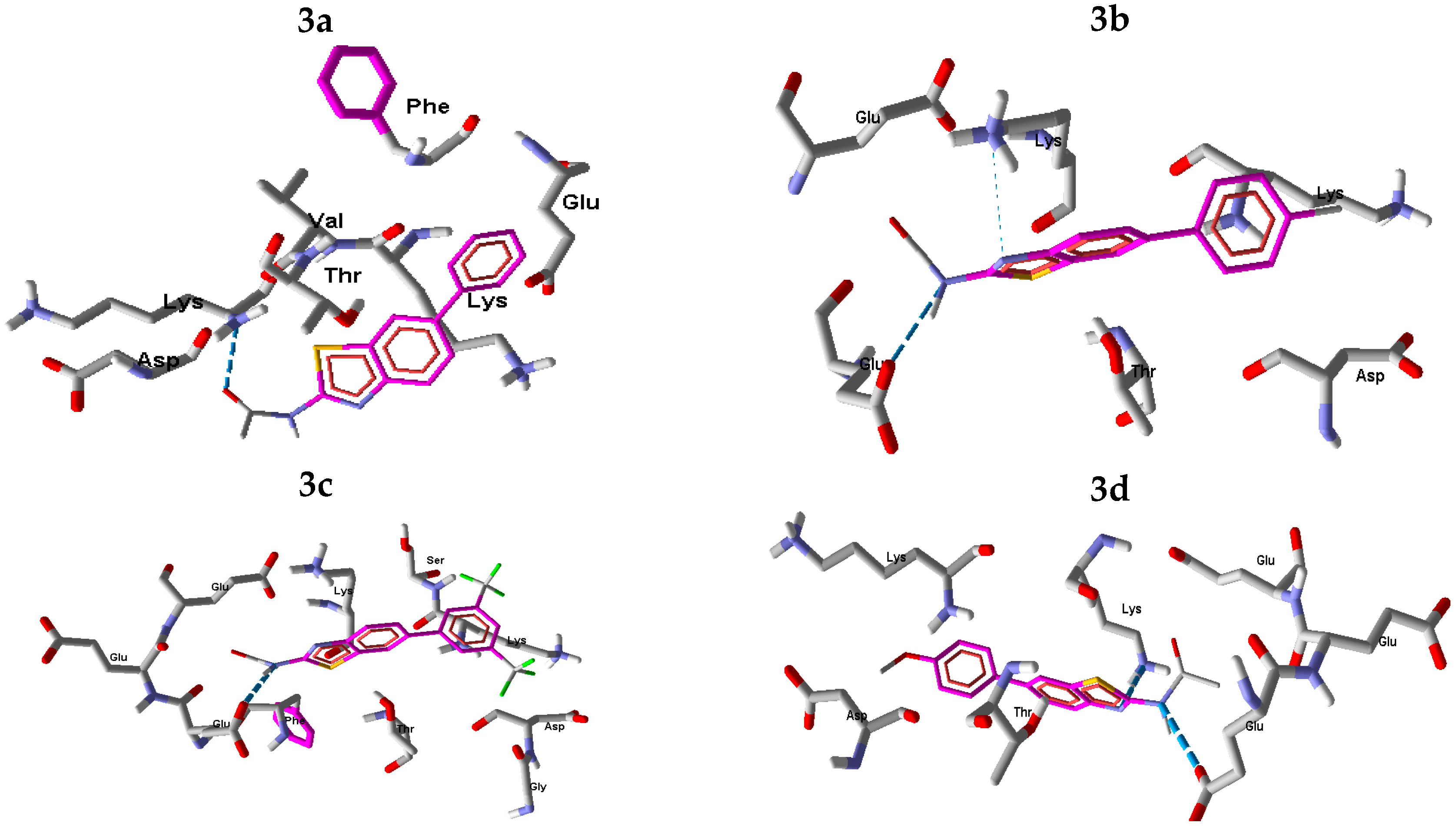
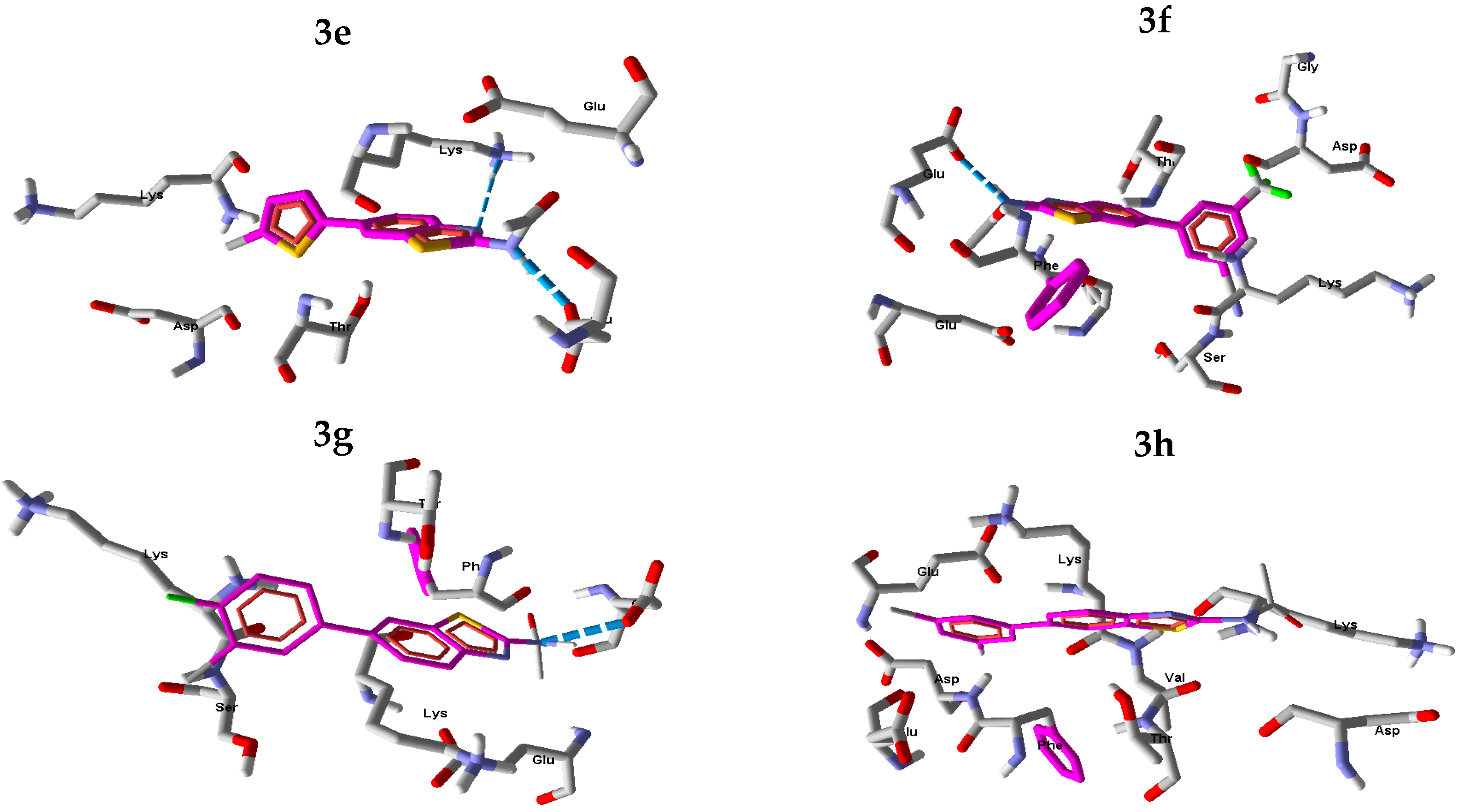
2.2.2. In Silico Studies with Urease
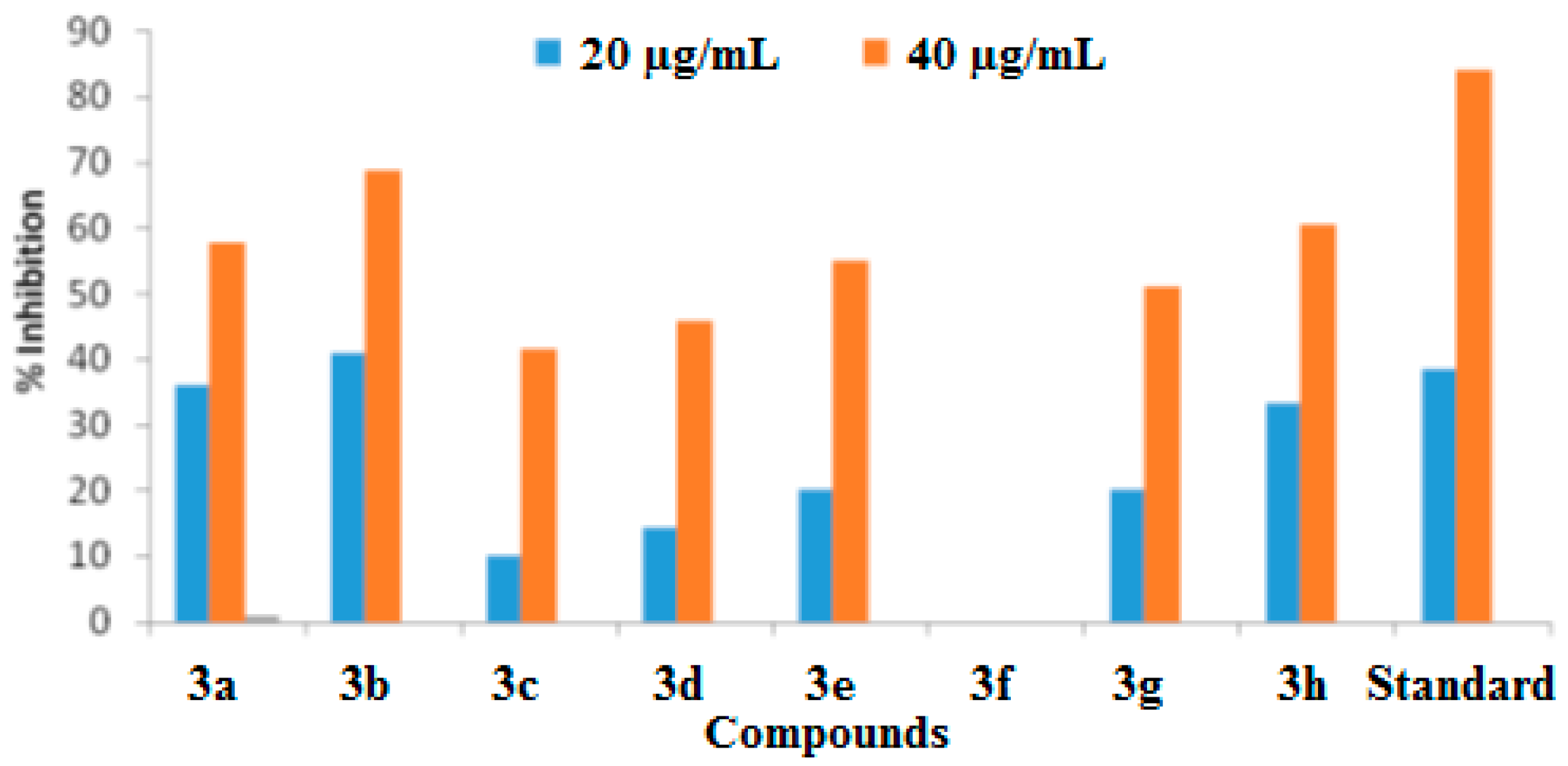
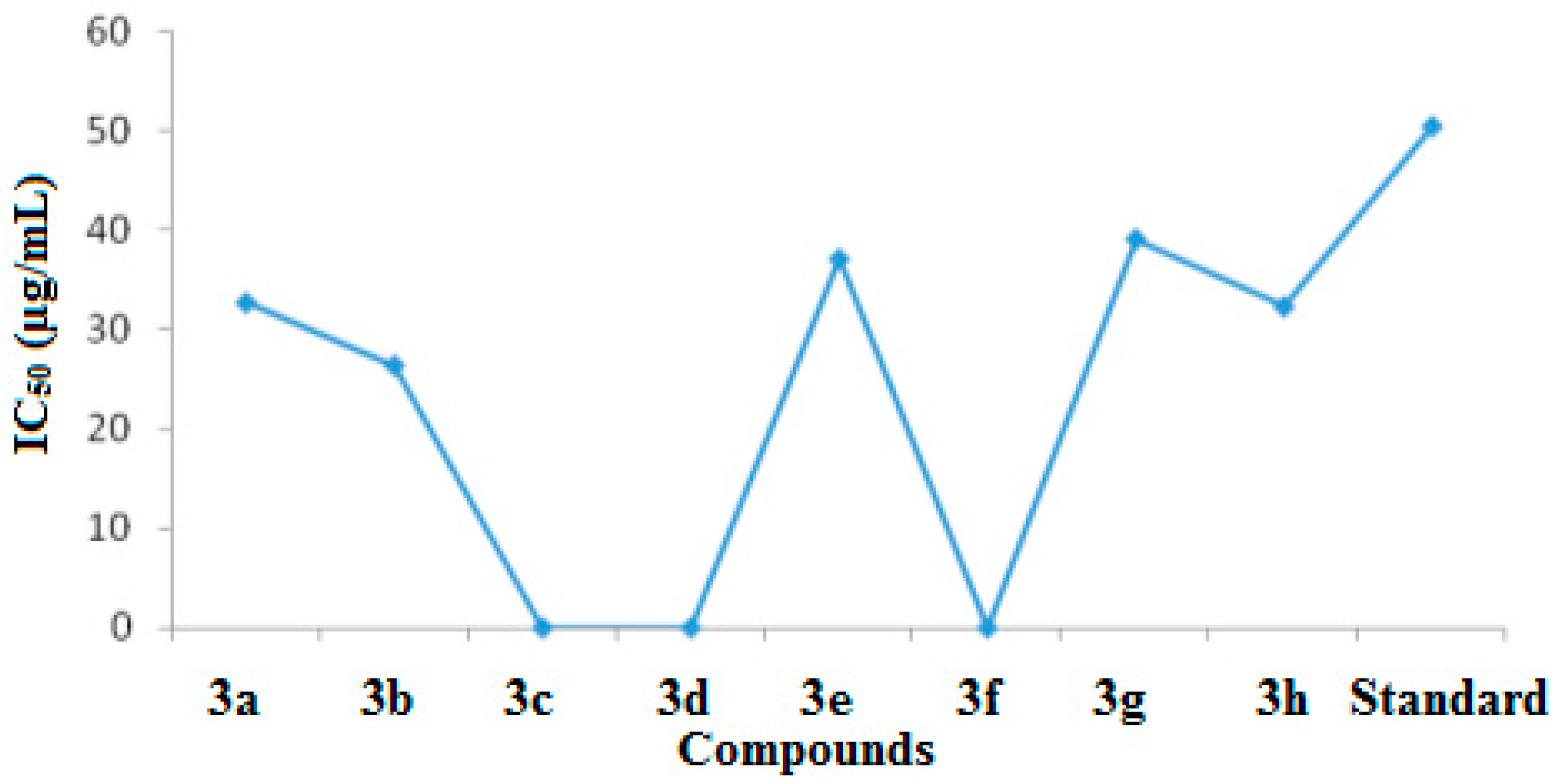
2.2.3. Nitric Oxide Scavenging Percentage Assay
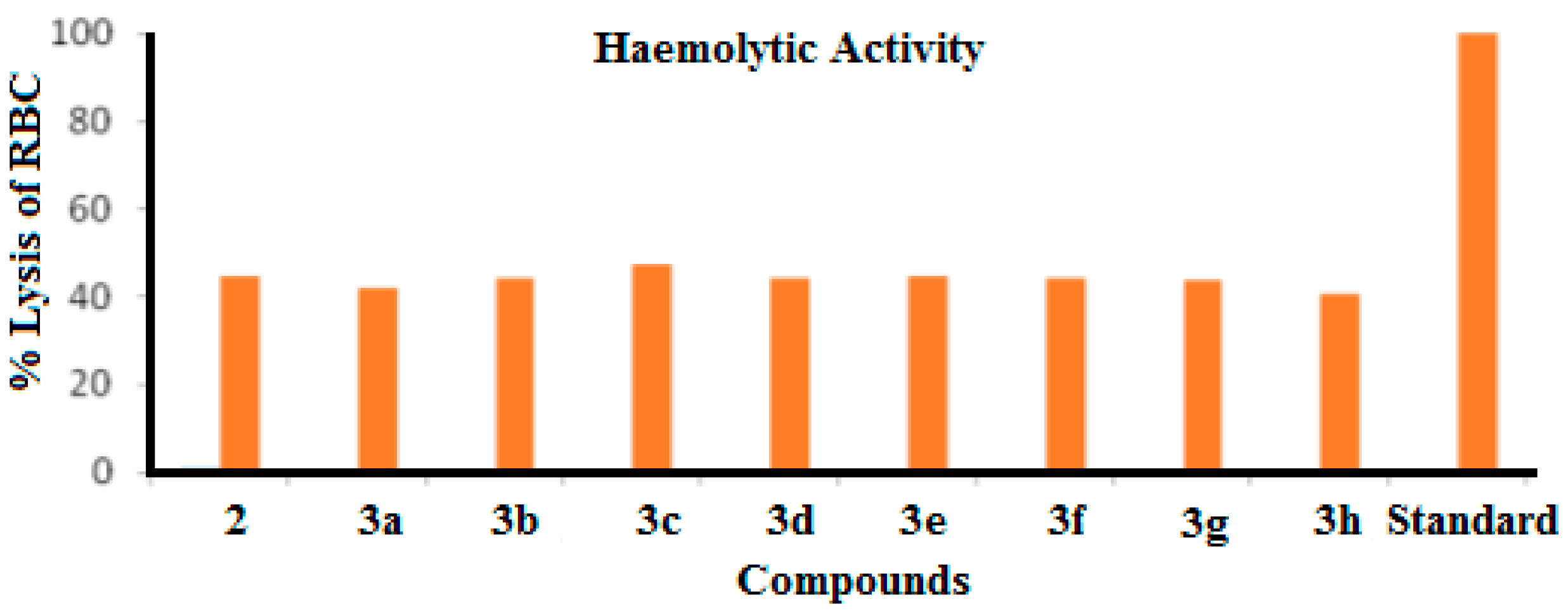
2.2.4. Haemolytic Activity
2.2.5. Antibacterial Activity
3. Experimental Section
3.1. General Information
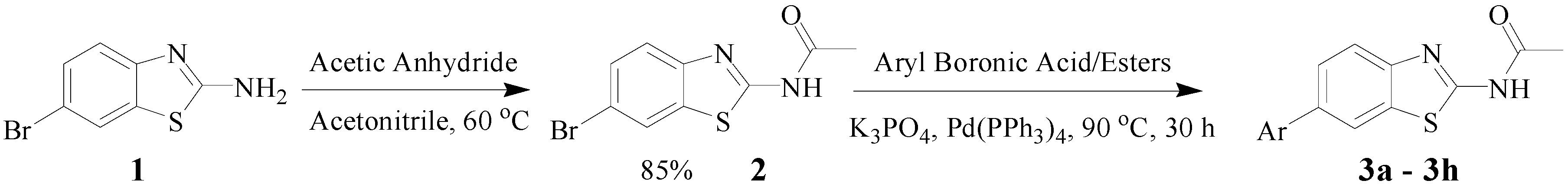
3.2. Procedure for the Preparation of N-(6-arylbenzo[d]thiazole-2-yl)acetamides 3a–3h
3.3. Procedure for Urease Inhibition Activity
3.4. Molecular Docking Study
3.5. Nitric Oxide Scavenging Activity
3.6. Haemolytic Activity
3.7. Antibacterial Activity
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gajdos, P.; Magdolen, P.; Zahradnik, P.; Foltinova, P. New conjugated benzothiazole-N-oxides: Synthesis and biological activity. Molecules 2009, 14, 5382–5388. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Behera, R.K.; Behera, P.K.; Mishra, B.K.; Behera, G.B. Cyanines during the 1990s: A review. Chem. Rev. 2000, 100, 1973–2012. [Google Scholar] [CrossRef] [PubMed]
- Michaelidou, A.S.; Hadjipavlou-Litina, D. Nonsteroidal anti-inflammatory drugs (NSAIDs): A comparative QSAR study. Chem. Rev. 2005, 105, 3235–3271. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.S.; Devprakash; Senthikumar, G.P. Benzothiazole: Different method of synthesis and diverse biological activities. Int. J. Pharm. Sci. Drug Res. 2011, 3, 1–7. [Google Scholar]
- Aiello, S.; Wells, G.; Stone, E.L.; Kadri, H.; Bazzi, R.; Bell, D.R.; Stevens, M.F.; Matthews, C.S.; Bradshaw, T.D.; Westwell, A.D. Synthesis and biological properties of benzothiazole, benzoxazole, and chromen-4-one analogues of the potent antitumor agent 2-(3,4-dimethoxyphenyl)-5-fluorobenzothiazole (PMX 610, NSC 721648). J. Med. Chem. 2008, 51, 5135–5139. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Ioerger, T.R.; Sacchettini, J.C. Discovery of novel nitrobenzothiazole inhibitors for mycobacterium tuberculosis ATP phosphoribosyl transferase (HisG) through virtual screening. J. Med. Chem. 2008, 51, 5984–5992. [Google Scholar] [CrossRef] [PubMed]
- Bondock, S.; Fadaly, W.; Metwally. Recent trends in the chemistry of 2-aminobenzothiazoles. J. Sulfur Chem. 2009, 30, 74–107. [Google Scholar] [CrossRef]
- Saeed, A.; Rafique, H.; Rasheed, S. Synthesis and antibacterial activity of some new 1-Aroyl-3-(substituated-2-benzothiazolyl)thioureas. Pharm. Chem. J. 2008, 48, 191–195. [Google Scholar] [CrossRef]
- Bele, D.S.; Singhvi, J. Synthesis of some mannich bases of 6-substituted-2-aminobenzothiazole as analgesic. Res. J. Pharm. Technol. 2014, 7, 316–321. [Google Scholar]
- Hutchinson, I.; Chua, M.; Bradshaw, T.D.; Matthews, C.S.; Stevens, F.G.; Westwell, A. Antitumor benzothiazoles, part 20: 3’-Cyano and 3’-alkynyl-substituted 2-(4’aminophenyl) benzothiazoles as new potent and selective analogues. Bioorg. Med. Chem. Lett. 2003, 13, 471–474. [Google Scholar] [CrossRef]
- Trapani, G.; Franco, M.; Latrofa, A.; Reho, A.; Liso, G. Synthesis in vitro and in vivo cytotoxicity, and prediction of the intestinal absorption of substituted 2-ethoxycarbonyl-imidazo[2,1-b]benzothiazoles. Eur. J. Pharm. Sci. 2001, 14, 209–216. [Google Scholar] [CrossRef]
- Kamal, M.; Dawood. Microwave-assisted Suzuki-Miyaura and Heck-Mizoroki cross-coupling reactions of aryl chlorides and bromides in water using stable benzothiazole-based palladium(II) precatalysts. Tetrahedron 2007, 63, 9642–9651. [Google Scholar]
- Ali, S.; Rasool, N.; Ullah, A.; Nasim, F.; Yaqoob, A.; Zubair, M.; Rashid, U.; Riaz, M. Design and synthesis of arylthiophene-2-Carbaldehydes via suzuki-miyaura reactions and their biological evaluation. Molecules 2013, 18, 14711–14725. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, K.; Zubair, M.; Rasool, N.; Ali, S.; Zahoor, A.F.; Rana, U.A.; Khan, S.; Shahid, M.; Zia-Ul-Haq, M.; Jaafar, H.Z. Regioselective synthesis of 2-(bromomethyl)-5-aryl-thiophene derivatives via palladium (0) catalyzed suzuki cross-coupling reactions: As antithrombotic and haemolytically active molecules. Chem. Cent. J. 2014, 8. [Google Scholar] [CrossRef]
- Noreen, M.; Rasool, N.; Khatib, M.E.; Molander, G.A. Arylation and heteroarylation of thienylsulfonamides with organotrifluoroborates. J. Org. Chem. 2014, 79, 7243–7249. [Google Scholar] [CrossRef] [PubMed]
- Ikram, H.M.; Rasool, N.; Ahmad, G.; Chotana, G.A.; Musharraf, S.G.; Zubair, M.; Rana, U.A.; Zia-Ul-Haq, M.; Jaafar, H.Z. Selective C-arylation of 2,5-dibromo-3-hexylthiophene via suzuki cross coupling reaction and their pharmacological aspects. Molecules 2015, 20, 5202–5214. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, T.; Rasool, N.; Noreen, M.; Gull, Y.; Zubair, M.; Ullah, A.; Rana, U.A. Palladium (0) catalyzed Suzuki cross-coupling reactions of 2,4-dibromothiophene: Selectivity, characterization and biological applications. J. Sulfur Chem. 2015, 36, 240–250. [Google Scholar] [CrossRef]
- Majo, V.J.; Prabhakaran, J.; Mann, J.J.; Kumar, J.S.D. An efficient palladium catalyzed synthesis of 2-arylbenzothiazoles. Tetrahedron Lett. 2003, 44, 8535–8537. [Google Scholar] [CrossRef]
- Gull, Y.; Rasool, N.; Noreen, M.; Nasim, F.; Yaqoob, A.; Kousar, S.; Rashid, U.; Bukhari, I.H.; Zubair, M.; Islam, M.S. Efficient synthesis of 2-amino-6-Arylbenzothiazoles via Pd(0) suzuki cross coupling reactions: Potent urease enzyme inhibition and nitric oxide scavenging activities of the products. Molecules 2013, 18, 8845–8857. [Google Scholar] [CrossRef] [PubMed]
- Benini, S.; Rypniewski, W.R.; Wilson, K.S.; Miletti, S.; Ciurli, S.; Mangani, S. A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: Why urea hydrolysis costs two nickels. Structure 1999, 7, 205–216. [Google Scholar] [CrossRef]
- Frishman, D.; Argos, P. Knowledge-based protein secondary structure assignment. Prot. Struct. Funct. Bioinform. 1995, 23, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Jamil, M.; Zubair, M.; Farid, M.A.; Altaf, A.A.; Rasool, N.; Nasim, F.U.H.; Ashraf, M.; Rashid, M.A.; Ejaz, S.A.; Yaqoob, A.; et al. Study of Antioxidant, Cytotoxic, and Enzyme Inhibition Activities of Some Symmetrical N3,N3′-Bis(disubstituted)isophthalyl-bis(thioureas) and N3,N3,N3′,N3′-Tetrakis(disubstituted)isophthalyl-bis(thioureas) and Their Cu(II) and Ni(II) Complexes. J. Chem. Soc. Pak. 2014, 36, 491–497. [Google Scholar]
- Jamil, M.; Zubair, M.; Altaf, A.A.; Farid, M.A.; Hussain, M.T.; Rasool, N.; Bukhari, I.H.; Ahmad, V.U. Synthesis, Characterization and Antibacterial Activity of Some Novel Symmetrical N-3,N-3′-Bis (disubstituted) isophthalyl-bis (thioureas) and N-3,N-3,N-3′,N-3′-Tetrakis (disubstituted) isophthalyl-bis (thiourea) and Their Cu (II) and Ni (II) Complexes. J. Chem. Soc. Pak. 2014, 35, 737–743. [Google Scholar]
- Onoda, Y.; Iwasaki, H.; Magaribuchi, T.; Tamaki, H. Effects of the new anti-ulcer agent 12-sulfodehydroabietic acid monosodium salt on healing of acetic acid-induced gastric ulcers in rats. Arzneim. Drug Res. 1991, 41, 546–548. [Google Scholar]
- Serwar, M.; Akhtar, T.; Hameed, S.; Khan, K.M. Synthesis urease inhibition and antimicrobial activities of some chiral 5-aryle-4-(1-phenylpropyl)-2H-1,2,4-trizole-3(4H)- thiones. ARKIVOC 2009, 7, 210–221. [Google Scholar]
- Mao, W.J.; Lv, P.C.; Shi, L.; Li, H.Q.; Zhu, H.L. Synthesis, molecular docking and biological evaluation of metronidazole derivatives as potent Helicobacter pylori urease inhibitors. Bioorg. Med. Chem. 2009, 17, 7531–7536. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.A.S.; Mahmood, S.U.; Shahid, M.; Saeed, A.; Iqbal, J. Synthesis, biological assay in vitro and molecular docking studies of new schiff base derivatives as potential urease inhibitors. Eur. J. Med. Chem. 2011, 46, 5473–5479. [Google Scholar] [CrossRef] [PubMed]
- Gul, R.; Rauf, M.; Badshah, A.; Azam, S.S.; Tahir, M.N.; Khan, A. Ferrocene-based guanidine derivatives: In vitro antimicrobial, DNA binding and docking supported urease inhibition studies. Eur. J. Med. Chem. 2014, 85, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Naresh, P.; Pattanaik, P.; Rajeshwar, B. Synthetic characterization and antioxidant screening of some novel 6-fluorobenzothiazole substituted [1,2,4] triazole analogues. Int. J. Pharm. Sci. 2013, 3, 170–174. [Google Scholar]
- Hazra, K.; Nargund, L.; Rashmi, P.; Chandra, J.N.S.; Nandha, B. Synthesis and antioxidant activity of some novel fluorobenzothiazolopyrazoline. Der. Chem. Sin. 2011, 2, 149–157. [Google Scholar]
- Devmurari, V.P.; Shivan, P.; Goyani, M.B.; Jivani, N.P. Synthesis and anticancer activity of some novel 2-substituared benzothiazole. Int. J. Chem. Sci. 2010, 8, 663–675. [Google Scholar]
- Powell, W.A.; Catranis, C.M.; Maynard, C.A. Design of self-processing antimicrobial peptides for plant protection. Lett. Appl. Microbiol. 2000, 31, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Saeed, S.; Rashid, N.; Jones, P.G.; Ali, M.; Hussain, R. Synthesis, characterization and biological evaluation of some thiourea derivatives bearing benzothiazole moiety as potential antimicrobial and anticancer agents. Eur. J. Med. Chem. 2010, 45, 1323–1331. [Google Scholar] [CrossRef] [PubMed]
- Miyaura, N.; Suzuki. Palladium-catalyzed cross-coupling reactions of organoboron compounds. Chem. Rev. 1995, 95, 2457–2483. [Google Scholar] [CrossRef]
- Pervez, H.; Rmzan, M.; Yaqub, M.; Nasim, F.H.; Khan, K. Synthesis and biological evaluation of some new N-4-aryl substituted 5-chloroisatin-3-thiosemicarbazones. Med. Chem. 2012, 8, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, A.; Ponnuraj, K. Crystal structure of the first plant urease from jack bean: 83 Years of journey from its first crystal to molecular structure. J. Mol.Biol. 2010, 400, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Rose, G.D. A physical basis for protein secondary structure. Proc. Nat. Acad. Sci. USA 1999, 96, 14258–14263. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; Mccammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Nat. Acad. Sci. USA 2010, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.; Olson, A.J.; Goodsell, D.S. Automated prediction of ligand-binding sites in proteins. Struct. Funct. Bioinform. 2003, 70, 1506–1517. [Google Scholar] [CrossRef] [PubMed]
- ChemAxon. Available online: http://www.chemaxon.com (accessed on 31 December 2015).
- CLC Drug Discovery Workbench. Available online: http://www.molegro.com/mmv-product.php (accessed on 31 December 2015).
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Mod. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Garrat, D. The Quantitative Analysis of Drugs, 3rd ed.; Chapman and Hall Ltd. Tokyo: Tokyo, Japan, 1996; pp. 456–458. [Google Scholar]
- Riaz, M.; Rasool, N.; Bukhari, I.H.; Shahid, M.; Zubair, M.; Rizwan, K.; Rashid, U. In vitro antimicrobial, antioxidant, cytotoxicity and GC-MS analysis of Mazus goodenifolius. Molecules 2011, 17, 14275–14287. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.U.; Awais, U.R.; Muhammad, A.B.; Hira, K.; Parsa, D. Synthesis and biological screening of N-substituted derivatives of N-benzyl-4-chlorobenzenesulfonamide. Asian J. Pharm. Health Sci. 2012, 2, 384–389. [Google Scholar]
- Sample Availability: Samples of all the newly synthesized compounds are available from the authors.


| Entry | Arylboronic Pinacol Ester/Arylboronic Acid | Product | H2O/Solvent (1:4) | Yields % |
|---|---|---|---|---|
| 1 |  | 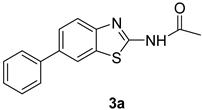 | Toluene 1,4-Dioxane | 75 80 |
| 2 |  | 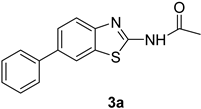 | 1,4-Dioxane | 77 |
| 3 | 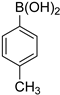 | 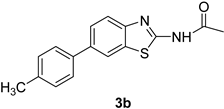 | 1,4-Dioxane | 85 |
| 4 |  | 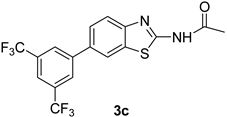 | 1,4-Dioxane | 81 |
| 5 |  | 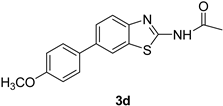 | 1,4-Dioxane | 79 |
| 6 |  |  | 1,4-Dioxane | 75 |
| 7 | 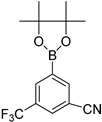 | 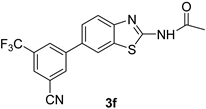 | 1,4-Dioxane | 77 |
| 8 |  | 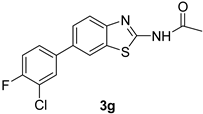 | 1,4-Dioxane | 79 |
| 9 | 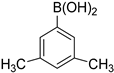 | 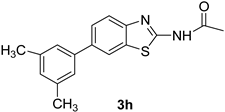 | 1,4-Dioxane | 83 |
| Compound | % Inhibition at 15 µg/mL | % Inhibition at 40 µg/mL | IC50 (µg/mL) |
|---|---|---|---|
| 3a | 44 ± 0.12 | 83.9 ± 0.12 | 18.6 |
| 3b | 47 ± 0.11 | 90.51 ± 0.19 | 16.5 |
| 3c | 46.09 ± 0.10 | 90.07 ± 0.20 | 17.2 |
| 3d | 46.5 ± 0.15 | 88.5 ± 0.24 | 17 |
| 3e | 46 ± 0.12 | 87 ± 0.26 | 17.2 |
| 3f | 42 ± 0.12 | 87.5 ± 0.26 | 19.2 |
| 3g | 42 ± 0.14 | 87 ± 0.26 | 18.9 |
| 3h | 43.59 ± 0.13 | 90.33 ± 0.20 | 18.4 |
| Standard | 47 ± 0.31 | 65 ± 0.01 | 23.1 |
| Compound | * IC50 (μg/mL) | Inhibition Constant | Binding Energy | Moldock Score |
|---|---|---|---|---|
| 3a | 18.6 | 267.93 | −4.87 | −83.39 |
| 3b | 16.5 | 232.29 | −4.96 | −81.68 |
| 3c | 17.2 | 232.56 | −4.96 | −68.81 |
| 3d | 17.0 | 434.76 | −4.59 | −69.20 |
| 3e | 17.2 | 278.10 | −4.85 | −87.00 |
| 3f | 19.2 | 90.570 | −5.52 | −87.90 |
| 3g | 18.9 | 153.46 | −5.20 | −79.91 |
| 3h | 18.4 | 150.67 | −5.21 | −80.11 |
| Compound | Number of H-bonds | H-Bonding Type (K--H--L) * | H-Bond Distance (K–L) (Å) | IC50 (μg/mL) | H-Binding Energy |
|---|---|---|---|---|---|
| 3a | 1 | O—H–N | 2.957 | 18.6 | −2.50 |
| 3b | 2 | N—H–N N–H—O | 2.837 3.544 | 16.5 | −3.45 |
| 3c | 1 | N–H—O | 2.782 | 17.2 | −2.98 |
| 3d | 2 | N–H—O N—H–N | 3.023 3.23 | 17.0 | −3.30 |
| 3e | 2 | N–H—O N—H–N | 2.906 3.388 | 17.2 | −3.01 |
| 3f | 1 | N–H—O | 2.739 | 19.2 | −1.67 |
| 3g | 1 | N–H—O | 2.696 | 18.9 | −1.99 |
| 3h | zero | -- | -- | 18.4 | -- |
| Compound | % Activity at 20 µg/mL | % Activity at 40 µg/mL | IC50 (µg/mL) |
|---|---|---|---|
| 3a | 36.25 ± 0.12 | 57.75 ± 0.12 | 32.7 |
| 3b | 41 ± 0.11 | 69 ± 0.12 | 26.4 |
| 3c | 10 ± 0.18 | 41.75 ± 0.20 | NC |
| 3d | 14.25 ± 0.17 | 46 ± 0.2 | NC |
| 3e | 20.25 ± 0.15 | 55 ± 0.31 | 37.1 |
| 3f | 0 | 0 | NC |
| 3g | 20.25 ± 0.15 | 51.25 ± 0.15 | 39.1 |
| 3h | 33.5 ± 0.13 | 60.5 ± 0.1 | 32.3 |
| Standard | 38.5 ± 0.16 | 84.1 ± 0.12 | 50.43 |
| Entry | % lysis of RBC | Entry | % lysis of RBC |
|---|---|---|---|
| 2 | 44.628 ± 0.369 | 3e | 44.425 ± 0.181 |
| 3a | 42.123 ± 0.479 | 3f | 44.063 ± 0.314 |
| 3b | 44.179 ± 0.157 | 3g | 43.614 ± 0.157 |
| 3c | 47.089 ± 0.130 | 3h | 40.661 ± 0.216 |
| 3d | 44.078 ± 0.279 | ||
| Standard | 99.78 ± 0.912 | ||
| Entry | % Activity at 40 μg/mL | |||||
|---|---|---|---|---|---|---|
| B. subtilis | S. aureus | P. aeruginosa | S. dysenteriae | S. typhae | E. coli | |
| 3a | - | - | - | 0.94 ± 0.45 | - | 37.52 ± 0.38 |
| 3b | - | - | - | - | - | 34.04 ± 0.40 |
| 3c | - | - | - | 0 ± 0.45 | 6.0 ± 0.47 | 49.05 ± 0.32 |
| 3d | - | - | - | 2.92 ± 0.44 | - | 42.10 ± 0.36 |
| 3e | - | - | - | - | - | 37.82 ± 0.38 |
| 3f | - | - | 7.54 ± 0.6 | 2.59 ± 0.44 | 1.2 ± 0.50 | 33.53 ± 0.41 |
| 3g | - | - | - | 1.45 ± 0.44 | - | 45.93 ± 0.3 |
| 3h | - | - | - | 3.51 ± o.43 | - | 22.89 ± 0.47 |
| Ampicillin | 23 ± 0.1 | 29 ± 0.61 | 25 ± 0.12 | 35 ± 0.32 | 29 ± 0.61 | 19 ± 0.31 |
| Entry | % Activity at 80 μg/mL | |||||
|---|---|---|---|---|---|---|
| B. subtilis | S. aureus | P. aeruginosa | S. dysenteriae | S. typhae | E. coli | |
| 3a | 6.08 ± 0.571 | 15.25 ± 0.5 | - | 8.47 ± 0.44 | 18.5 ± 0.58 | 57.97 ± 0.25 |
| 3b | - | - | - | 6.57 ± 0.45 | 17.7 ± 0.5 | 56.13 ± 0.32 |
| 3c | - | - | - | - | 26.2 ± 0.53 | 50.49 ± 0.30 |
| 3d | 7.31 ± 0.5635 | - | - | 5.92 ± 0.46 | 28.86 ± 0.5 | 51 ± 0.30 |
| 3e | 12.66 ± 0.531 | 3.75 ± 0.65 | - | - | 19.2 ± 0.5 | 50.8 ± 0.30 |
| 3f | 4.27 ± 0.582 | 8.89 ± 0.5 | 2.96 ± 0.6 | 10.93 ± 0.43 | 25 ± 0.54 | 55 ± 0.32 |
| 3g | - | - | - | 11.51 ± 0.42 | 21.05 ± 0.5 | 57.84 ± 0.25 |
| 3h | 5.67 ± 0.5735 | - | - | 7.48 ± 0.45 | 17.43 ± 0.5 | 53.96 ± 0.40 |
| Ampicillin | 50.5 ± 0.31 | 52.9 ± 0.29 | 52 ± 0.26 | 56 ± 0.26 | 42.9 ± 0.29 | 45.9 ± 0.21 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gull, Y.; Rasool, N.; Noreen, M.; Altaf, A.A.; Musharraf, S.G.; Zubair, M.; Nasim, F.-U.-H.; Yaqoob, A.; DeFeo, V.; Zia-Ul-Haq, M. Synthesis of N-(6-Arylbenzo[d]thiazole-2-acetamide Derivatives and Their Biological Activities: An Experimental and Computational Approach. Molecules 2016, 21, 266. https://doi.org/10.3390/molecules21030266
Gull Y, Rasool N, Noreen M, Altaf AA, Musharraf SG, Zubair M, Nasim F-U-H, Yaqoob A, DeFeo V, Zia-Ul-Haq M. Synthesis of N-(6-Arylbenzo[d]thiazole-2-acetamide Derivatives and Their Biological Activities: An Experimental and Computational Approach. Molecules. 2016; 21(3):266. https://doi.org/10.3390/molecules21030266
Chicago/Turabian StyleGull, Yasmeen, Nasir Rasool, Mnaza Noreen, Ataf Ali Altaf, Syed Ghulam Musharraf, Muhammad Zubair, Faiz-Ul-Hassan Nasim, Asma Yaqoob, Vincenzo DeFeo, and Muhammad Zia-Ul-Haq. 2016. "Synthesis of N-(6-Arylbenzo[d]thiazole-2-acetamide Derivatives and Their Biological Activities: An Experimental and Computational Approach" Molecules 21, no. 3: 266. https://doi.org/10.3390/molecules21030266