Gold Nanoparticles in Single-Cell Analysis for Surface Enhanced Raman Scattering
Abstract
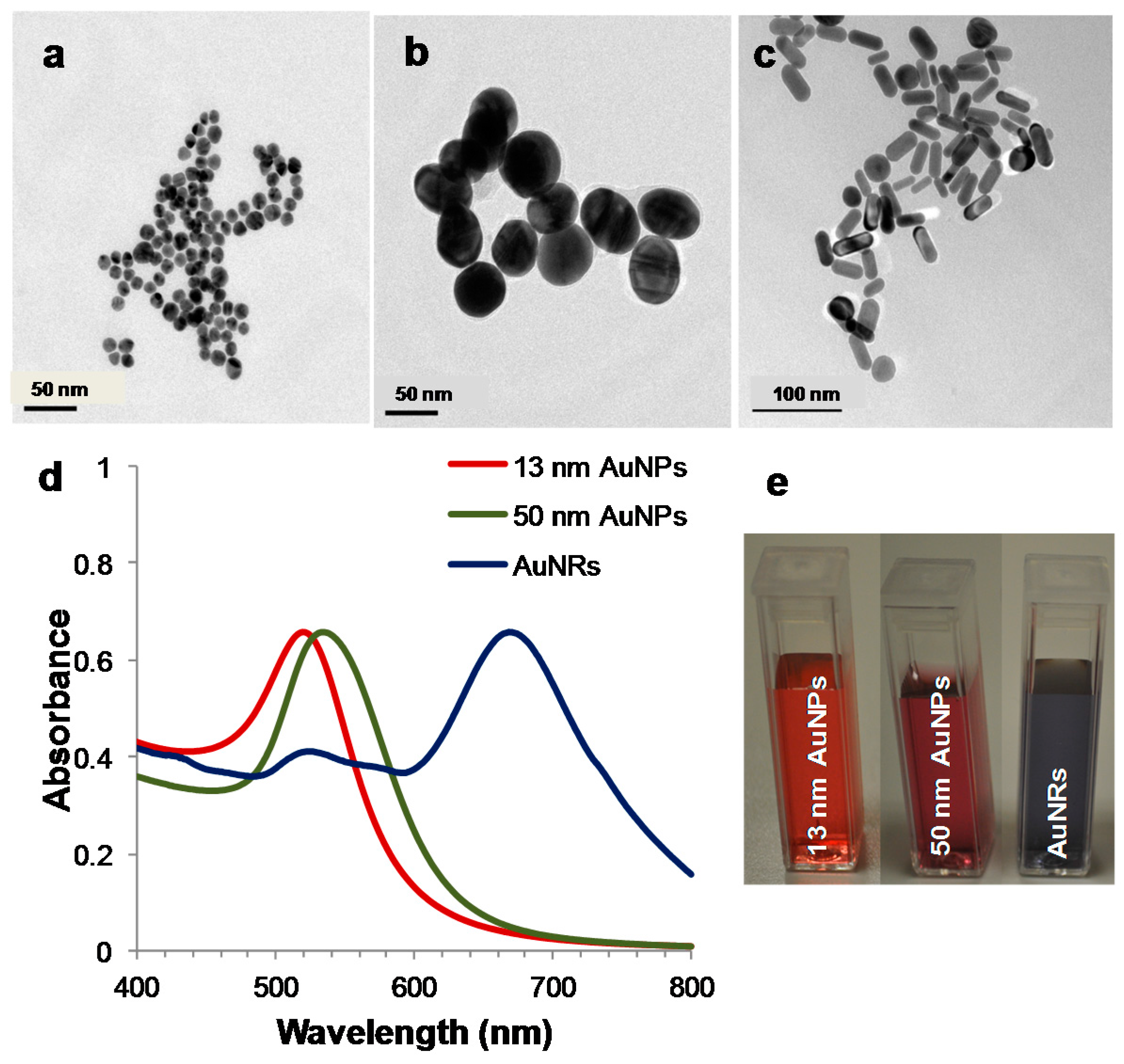
:1. Introduction
2. Cellular Interaction and Toxicity Concerns of Gold Nanoparticles
2.1. Cellular Interaction and Uptake of AuNPs
2.2. Influence of Size, Shape and Surface Chemistry on AuNP Toxicity
3. Single-Cell Analysis and Surface-Enhanced Raman Scattering
3.1. Single-Cell Analysis
3.2. Raman Spectroscopy and Surface-Enhanced Raman Scattering
3.3. SERS Substrates Used in Biological Applications
3.4. SERS-Based Single-Cell Studies
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Lee, K.-S.; El-Sayed, M.A. Dependence of the enhanced optical scattering efficiency relative to that of absorption for gold metal nanorods on aspect ratio, size, end-cap shape, and medium refractive index. J. Phys. Chem. B 2005, 109, 20331–20338. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.K.; Pal, T. Interparticle coupling effect on the surface plasmon resonance of gold nanoparticles: From theory to applications. Chem. Rev. 2007, 107, 4797–4862. [Google Scholar] [CrossRef] [PubMed]
- Willets, K.A.; Van Duyne, R.P. Localized surface plasmon resonance spectroscopy and sensing. Annu. Rev. Phys. Chem. 2007, 58, 267–297. [Google Scholar] [CrossRef] [PubMed]
- Ozbay, E. Plasmonics: Merging photonics and electronics at nanoscale dimensions. Science 2006, 311, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Daniel, M.-C.; Astruc, D. Gold nanoparticles: Assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem. Rev. 2004, 104, 293–346. [Google Scholar] [CrossRef] [PubMed]
- Saha, K.; Agasti, S.S.; Kim, C.; Li, X.; Rotello, V.M. Gold nanoparticles in chemical and biological sensing. Chem. Rev. 2012, 112, 2739–2779. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-J.; Liu, Y.-G.; Jiang, L.-P.; Zhu, J.-J. Synthesis, characterizations of silica-coated gold nanorods and its applications in electroanalysis of hemoglobin. Electrochem. Commun. 2008, 10, 355–358. [Google Scholar] [CrossRef]
- Song, T.; Tang, L.; Tan, L.H.; Wang, X.; Satyavolu, N.S.R.; Xing, H.; Wang, Z.; Li, J.; Liang, H.; Lu, Y. DNA-encoded tuning of geometric and plasmonic properties of nanoparticles growing from gold nanorod seeds. Angew. Chem. Int. Ed. 2015, 54, 8114–8118. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.H.; Yue, Y.; Satyavolu, N.S.R.; Ali, A.S.; Wang, Z.; Wu, Y.; Lu, Y. Mechanistic insight into DNA-guided control of nanoparticle morphologies. J. Am. Chem. Soc. 2015, 137, 14456–14464. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-J.; Gu, M.-M.; Zheng, T.-T.; Zhu, J.-J. Synthesis of gelatin-stabilized gold nanoparticles and assembly of carboxylic single-walled carbon nanotubes/Au composites for cytosensing and drug uptake. Anal. Chem. 2009, 81, 6641–6648. [Google Scholar] [CrossRef] [PubMed]
- Busbee, B.D.; Obare, S.O.; Murphy, C.J. An improved synthesis of high-aspect-ratio gold nanorods. Adv. Mater. 2003, 15, 414–416. [Google Scholar] [CrossRef]
- Nikoobakht, B.; El-Sayed, M.A. Preparation and growth mechanism of gold nanorods (NRS) using seed-mediated growth method. Chem. Mater. 2003, 15, 1957–1962. [Google Scholar] [CrossRef]
- Alkilany, A.M.; Nagaria, P.K.; Hexel, C.R.; Shaw, T.J.; Murphy, C.J.; Wyatt, M.D. Cellular uptake and cytotoxicity of gold nanorods: Molecular origin of cytotoxicity and surface effects. Small 2009, 5, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Huang, X.; Liu, H.; Zan, F.; Ren, J. Colloidal stability of gold nanoparticles modified with thiol compounds: Bioconjugation and application in cancer cell imaging. Langmuir 2012, 28, 4464–4471. [Google Scholar] [CrossRef] [PubMed]
- Aioub, M.; El-Sayed, M.A. A real-time surface enhanced raman spectroscopy study of plasmonic photothermal cell death using targeted gold nanoparticles. J. Am. Chem. Soc. 2016, 138, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Austin, L.A.; Kang, B.; El-Sayed, M.A. Probing molecular cell event dynamics at the single-cell level with targeted plasmonic gold nanoparticles: A review. Nano Today 2015, 10, 542–558. [Google Scholar] [CrossRef]
- Kang, J.W.; So, P.T.; Dasari, R.R.; Lim, D.-K. High resolution live cell raman imaging using subcellular organelle-targeting sers-sensitive gold nanoparticles with highly narrow intra-nanogap. Nano Lett. 2015, 15, 1766–1772. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Singh, A.K.; Khan, S.A.; Senapati, D.; Yu, H.; Ray, P.C. Gold nano-popcorn-based targeted diagnosis, nanotherapy treatment, and in situ monitoring of photothermal therapy response of prostate cancer cells using surface-enhanced raman spectroscopy. J. Am. Chem. Soc. 2010, 132, 18103–18114. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Bai, X.; Su, L.; Du, Z.; Shen, A.; Materny, A.; Hu, J. Combined labelled and label-free sers probes for triplex three-dimensional cellular imaging. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gao, H.; Bao, G. Physical principles of nanoparticle cellular endocytosis. ACS Nano 2015, 9, 8655–8671. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhao, Y.; Liu, Y.; Chang, X.; Chen, C.; Zhao, Y. Cellular uptake, intracellular trafficking, and cytotoxicity of nanomaterials. Small 2011, 7, 1322–1337. [Google Scholar] [CrossRef] [PubMed]
- Yameen, B.; Choi, W.I.; Vilos, C.; Swami, A.; Shi, J.; Farokhzad, O.C. Insight into nanoparticle cellular uptake and intracellular targeting. J. Control. Release 2014, 190, 485–499. [Google Scholar] [CrossRef] [PubMed]
- Champion, J.A.; Mitragotri, S. Shape induced inhibition of phagocytosis of polymer particles. Pharm. Res. 2009, 26, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Choi, I.-H. Phagocytosis and endocytosis of silver nanoparticles induce interleukin-8 production in human macrophages. Yonsei Med. J. 2012, 53, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Bansal, V.; Chaudhary, M.; Basu, A.; Bhonde, R.R.; Sastry, M. Biocompatibility of gold nanoparticles and their endocytotic fate inside the cellular compartment: A microscopic overview. Langmuir 2005, 21, 10644–10654. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Yang, Z.; Zhang, S.; Cao, S.; Shen, S.; Pang, Z.; Jiang, X. Ligand modified nanoparticles increases cell uptake, alters endocytosis and elevates glioma distribution and internalization. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Zhou, H.; Liu, H.; Gao, J. The cytotoxicity of gold nanoparticles is dispersity-dependent. Dalton Trans. 2015, 44, 17911–17915. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.T.; Tang, F.M.A.; Li, J.J.E.; Ong, C.; Yung, L.L.Y.; Bay, B.H. Clathrin-mediated endocytosis of gold nanoparticles in vitro. Anat. Rec. 2015, 298, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Contreras, J.; Xie, J.; Chen, Y.J.; Pei, H.; Zhang, G.; Fraser, C.L.; Hamm-Alvarez, S.F. Intracellular uptake and trafficking of difluoroboron dibenzoylmethane-polylactide nanoparticles in hela cells. ACS Nano 2010, 4, 2735–2747. [Google Scholar] [CrossRef] [PubMed]
- Partlow, K.C.; Lanza, G.M.; Wickline, S.A. Exploiting lipid raft transport with membrane targeted nanoparticles: A strategy for cytosolic drug delivery. Biomaterials 2008, 29, 3367–3375. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hu, X.; Li, J.; Russe, A.C.M.; Kawazoe, N.; Yang, Y.; Chen, G. Influence of cell size on cellular uptake of gold nanoparticles. Biomater. Sci. 2016, 4, 970–978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-D.; Wu, D.; Shen, X.; Liu, P.-X.; Yang, N.; Zhao, B.; Zhang, H.; Sun, Y.-M.; Zhang, L.-A.; Fan, F.-Y. Size-dependent in vivo toxicity of peg-coated gold nanoparticles. Int. J. Nanomedicine 2011, 6, 2071–2081. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.A.; Salleng, K.J.; Cliffel, D.E.; Feldheim, D.L. In vivo toxicity, biodistribution, and clearance of glutathione-coated gold nanoparticles. Nanomedicine 2013, 9, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-D.; Wu, H.-Y.; Wu, D.; Wang, Y.-Y.; Chang, J.-H.; Zhai, Z.-B.; Meng, A.-M.; Liu, P.-X.; Zhang, L.-A.; Fan, F.-Y. Toxicologic effects of gold nanoparticles in vivo by different administration routes. Int. J. Nanomed. 2010, 5, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-D.; Wu, D.; Shen, X.; Liu, P.-X.; Fan, F.-Y.; Fan, S.-J. In vivo renal clearance, biodistribution, toxicity of gold nanoclusters. Biomaterials 2012, 33, 4628–4638. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Dorrigan, A.; Saad, S.; Hare, D.J.; Cortie, M.B.; Valenzuela, S.M. In vivo study of spherical gold nanoparticles: Inflammatory effects and distribution in mice. PLoS ONE 2013, 8, e58208. [Google Scholar] [CrossRef] [PubMed]
- Favi, P.M.; Gao, M.; Johana Sepúlveda Arango, L.; Ospina, S.P.; Morales, M.; Pavon, J.J.; Webster, T.J. Shape and surface effects on the cytotoxicity of nanoparticles: Gold nanospheres versus gold nanostars. J. Biomed. Mater. Res. Part A 2015, 103, 3449–3462. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, B.D.; Ghazani, A.A.; Chan, W.C. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006, 6, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; He, L.; McClements, D.J.; Xiao, H. Uptake of gold nanoparticles by intestinal epithelial cells: Impact of particle size on their absorption, accumulation, and toxicity. J. Agric. Food Chem. 2015, 63, 8044–8049. [Google Scholar] [CrossRef] [PubMed]
- Moser, F.; Hildenbrand, G.; Müller, P.; Al Saroori, A.; Biswas, A.; Bach, M.; Wenz, F.; Cremer, C.; Burger, N.; Veldwijk, M.R. Cellular uptake of gold nanoparticles and their behavior as labels for localization microscopy. Biophys. J. 2016, 110, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Zhang, Q.; Xu, H.; Yan, Z. Effects of nanoparticle size on antitumor activity of 10-hydroxycamptothecin-conjugated gold nanoparticles: In vitro and in vivo studies. Int. J. Nanomed. 2016, 11, 929–940. [Google Scholar]
- Jiang, Y.; Huo, S.; Mizuhara, T.; Das, R.; Lee, Y.-W.; Hou, S.; Moyano, D.F.; Duncan, B.; Liang, X.-J.; Rotello, V.M. The interplay of size and surface functionality on the cellular uptake of sub-10 nm gold nanoparticles. ACS Nano 2015, 9, 9986–9993. [Google Scholar] [CrossRef] [PubMed]
- Stojiljković, A.; Kuehni-Boghenbor, K.; Gaschen, V.; Schüpbach, G.; Mevissen, M.; Kinnear, C.; Möller, A.-M.; Stoffel, M. High-content analysis of factors affecting gold nanoparticle uptake by neuronal and microglial cells in culture. Nanoscale 2016, 8, 16650–16661. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-S.; Hung, Y.-C.; Liau, I.; Huang, G.S. Assessment of the in vivo toxicity of gold nanoparticles. Nanoscale Res. Lett. 2009, 4, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tang, S.; Wang, B.; Li, Y.; Huang, H.; Wang, H.; Li, P.; Li, C.; Chu, P.K.; Yu, X.-F. Metabolizable small gold nanorods: Size-dependent cytotoxicity, cell uptake and in vivo biodistribution. ACS Biomater. Sci. Eng. 2016, 2, 789–797. [Google Scholar] [CrossRef]
- Gao, B.; Xu, J.; Shen, L.; Chen, H.; Yang, H.-J.; Li, A.-H.; Xiao, W.-H. Cellular uptake and intra-organ biodistribution of functionalized silica-coated gold nanorods. Mol. Imaging Biol. 2016, 18, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Robinson, R.; Gerlach, W.; Ghandehari, H. Comparative effect of gold nanorods and nanocages for prostate tumor hyperthermia. J. Control. Release 2015, 220, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Tatini, F.; Landini, I.; Scaletti, F.; Massai, L.; Centi, S.; Ratto, F.; Nobili, S.; Romano, G.; Fusi, F.; Messori, L. Size dependent biological profiles of pegylated gold nanorods. J. Mater. Chem. B 2014, 2, 6072–6080. [Google Scholar] [CrossRef]
- Walling, M.A.; Shepard, J.R. Cellular heterogeneity and live cell arrays. Chem. Soc. Rev. 2011, 40, 4049–4076. [Google Scholar] [CrossRef] [PubMed]
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta Rev. Cancer 2010, 1805, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Perkins, T.J.; Swain, P.S. Strategies for cellular decision-making. Mol. Syst. Biol. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Irish, J.M.; Kotecha, N.; Nolan, G.P. Mapping normal and cancer cell signalling networks: Towards single-cell proteomics. Nat. Rev. Cancer 2006, 6, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Graf, T.; Stadtfeld, M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell 2008, 3, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Friedman, N.; Xie, X.S. Stochastic protein expression in individual cells at the single molecule level. Nature 2006, 440, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, N.; Young, J.W.; Alon, U.; Swain, P.S.; Elowitz, M.B. Gene regulation at the single-cell level. Science 2005, 307, 1962–1965. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, L.S.; Burnett, J.C.; Toettcher, J.E.; Arkin, A.P.; Schaffer, D.V. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 tat fluctuations drive phenotypic diversity. Cell 2005, 122, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Toriello, N.M.; Douglas, E.S.; Thaitrong, N.; Hsiao, S.C.; Francis, M.B.; Bertozzi, C.R.; Mathies, R.A. Integrated microfluidic bioprocessor for single-cell gene expression analysis. Proc. Natl. Acad. Sci. USA 2008, 105, 20173–20178. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wu, J.; Dressman, D.C.; Iacobuzio-Donahue, C.; Markowitz, S.D.; Velculescu, V.E.; Diaz, L.A., Jr.; Kinzler, K.W.; Vogelstein, B.; Papadopoulos, N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 2010, 464, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.R.; Ribas, A.; Mischel, P.S. Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug Discov. 2016, 15, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Bodovitz, S. Single cell analysis: The new frontier in ‘omics’. Trends Biotechnol. 2010, 28, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Amir, E.-a.D.; Davis, K.L.; Tadmor, M.D.; Simonds, E.F.; Levine, J.H.; Bendall, S.C.; Shenfeld, D.K.; Krishnaswamy, S.; Nolan, G.P.; Pe’er, D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013, 31, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Jaitin, D.A.; Kenigsberg, E.; Keren-Shaul, H.; Elefant, N.; Paul, F.; Zaretsky, I.; Mildner, A.; Cohen, N.; Jung, S.; Tanay, A. Massively parallel single-cell rna-seq for marker-free decomposition of tissues into cell types. Science 2014, 343, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Bruggner, R.V.; Bodenmiller, B.; Dill, D.L.; Tibshirani, R.J.; Nolan, G.P. Automated identification of stratifying signatures in cellular subpopulations. Proc. Natl. Acad. Sci. USA 2014, 111, E2770–E2777. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Xue, Q.; Eisele, M.R.; Sulistijo, E.S.; Brower, K.; Han, L.; Amir, E.-A.D.; Pe’er, D.; Miller-Jensen, K.; Fan, R. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc. Natl. Acad. Sci. USA 2015, 112, E607–E615. [Google Scholar] [CrossRef] [PubMed]
- Boiani, M.; Cibelli, J.B. What we can learn from single-cell analysis in development. Mol. Hum. Reprod. 2016, 22, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Prats Mateu, B.; Harreither, E.; Schosserer, M.; Puxbaum, V.; Gludovacz, E.; Borth, N.; Gierlinger, N.; Grillari, J. Label-free live cell imaging by confocal raman microscopy identifies cho host and producer cell lines. Biotechnol. J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.J.; Ahrens, M.B. Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron 2015, 85, 462–483. [Google Scholar] [CrossRef] [PubMed]
- Rajamohan, D.; Kalra, S.; Duc Hoang, M.; George, V.; Staniforth, A.; Russell, H.; Yang, X.; Denning, C. Automated electrophysiological and pharmacological evaluation of human pluripotent stem cell-derived cardiomyocytes. Stem Cells Dev. 2016, 25, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Sakmann, B.; Neher, E. Patch clamp techniques for studying ionic channels in excitable membranes. Annu. Rev. Physiol. 1984, 46, 455–472. [Google Scholar] [CrossRef] [PubMed]
- Davison, G. Exercise and oxidative damage in nucleoid DNA quantified using single cell gel electrophoresis: Present and future application. Front. Physiol. 2016, 7, 249. [Google Scholar] [CrossRef] [PubMed]
- Speit, G.; Hartmann, A. The comet assay (single-cell gel test). DNA Repair Protoc. Eukaryot. Syst. 1999, 113, 203–212. [Google Scholar]
- Samsel, L.; McCoy, J.P. Detection and characterization of rare circulating endothelial cells by imaging flow cytometry. Imaging Flow Cytom. Methods Protoc. 2016, 84, 249–264. [Google Scholar]
- Bintu, L.; Yong, J.; Antebi, Y.E.; McCue, K.; Kazuki, Y.; Uno, N.; Oshimura, M.; Elowitz, M.B. Dynamics of epigenetic regulation at the single-cell level. Science 2016, 351, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Lin, L.; Zhang, J.; He, Z.; Uchiyama, K.; Lin, J.-M. Single-cell analysis using drop-on-demand inkjet printing and probe electrospray ionization mass spectrometry. Anal. Chem. 2016, 88, 4354–4360. [Google Scholar] [CrossRef] [PubMed]
- Onjiko, R.M.; Morris, S.E.; Moody, S.A.; Nemes, P. Single-cell mass spectrometry with multi-solvent extraction identifies metabolic differences between left and right blastomeres in the 8-cell frog (xenopus) embryo. Analyst 2016, 141, 3648–3656. [Google Scholar] [CrossRef] [PubMed]
- McKelvey-Martin, V.; Green, M.; Schmezer, P.; Pool-Zobel, B.; De Meo, M.; Collins, A. The single cell gel electrophoresis assay (comet assay): A European review. Mutat. Res. 1993, 288, 47–63. [Google Scholar] [CrossRef]
- Vo-Dinh, T.; Kasili, P. Fiber-optic nanosensors for single-cell monitoring. Anal. Bioanal. Chem. 2005, 382, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Casey, D.; Dooley, J. Optical tools for single-cell manipulation and analysis. In Essentials of Single-Cell Analysis; Springer: New York, NY, USA, 2016; pp. 131–157. [Google Scholar]
- Hu, C.; Shen, J.; Yan, J.; Zhong, J.; Qin, W.; Liu, R.; Aldalbahi, A.; Zuo, X.; Song, S.; Fan, C. Highly narrow nanogap-containing Au@Au core–shell SERS nanoparticles: Size-dependent raman enhancement and applications in cancer cell imaging. Nanoscale 2016, 8, 2090–2096. [Google Scholar] [CrossRef] [PubMed]
- El-Said, W.A.; Kim, T.-H.; Kim, H.; Choi, J.-W. Analysis of intracellular state based on controlled 3d nanostructures mediated surface enhanced raman scattering. PLoS ONE 2011, 6, e15836. [Google Scholar] [CrossRef] [PubMed]
- Belov, L.; Zhou, J.; Christopherson, R.I. Cell surface markers in colorectal cancer prognosis. Int. J. Mol. Sci. 2010, 12, 78–113. [Google Scholar] [CrossRef] [PubMed]
- Xia, P. Surface markers of cancer stem cells in solid tumors. Curr. Stem Cell Res. Ther. 2014, 9, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Karsten, U.; Goletz, S. What makes cancer stem cell markers different? SpringerPlus 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Dieriks, B.; De Vos, W.H.; Moreels, M.; Ghardi, M.; Hennekam, R.; Broers, J.; Baatout, S.; Van Oostveldt, P. Multiplexed profiling of secreted proteins for the detection of potential space biomarkers. Mol. Med. Rep. 2011, 4, 17–23. [Google Scholar] [PubMed]
- Kuppusamy, P.; Govindan, N.; Yusoff, M.M.; Ichwan, S.J. Proteins are potent biomarkers to detect colon cancer progression. Saudi J. Biol. Sci. 2014. [Google Scholar] [CrossRef]
- Liao, M.-C.; Muratore, C.R.; Gierahn, T.M.; Sullivan, S.E.; Srikanth, P.; De Jager, P.L.; Love, J.C.; Young-Pearse, T.L. Single-cell detection of secreted aβ and sappα from human ipsc-derived neurons and astrocytes. J. Neurosci. 2016, 36, 1730–1746. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, A.C.; Miname, M.H.; Santos, R.D. Uric acid: A marker of increased cardiovascular risk. Atherosclerosis 2009, 202, 11–17. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, T.; Abbasi, F.; Cheal, K.; Chu, J.; Lamendola, C.; Reaven, G. Use of metabolic markers to identify overweight individuals who are insulin resistant. Ann. Intern. Med. 2003, 139, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Irish, J.M.; Hovland, R.; Krutzik, P.O.; Perez, O.D.; Bruserud, Ø.; Gjertsen, B.T.; Nolan, G.P. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell 2004, 118, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Shin, Y.S.; Xue, M.; Matsutani, T.; Masui, K.; Yang, H.; Ikegami, S.; Gu, Y.; Herrmann, K.; Johnson, D. Single-cell phosphoproteomics resolves adaptive signaling dynamics and informs targeted combination therapy in glioblastoma. Cancer Cell 2016, 29, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Kalisky, T.; Quake, S.R. Single-cell genomics. Nat. Methods 2011, 8, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, I.C.; Voet, T. Single cell genomics: Advances and future perspectives. PLoS Genet. 2014, 10, e1004126. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, R. Entering the era of single-cell transcriptomics in biology and medicine. Nat. Methods 2014, 11, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef] [PubMed]
- Perfetto, S.P.; Chattopadhyay, P.K.; Roederer, M. Seventeen-colour flow cytometry: Unravelling the immune system. Nat. Rev. Immunol. 2004, 4, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Saeys, Y.; Van Gassen, S.; Lambrecht, B.N. Computational flow cytometry: Helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol. 2016, 16, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Sendra, V.G.; Lie, A.; Romain, G.; Agarwal, S.K.; Varadarajan, N. Detection and isolation of auto-reactive human antibodies from primary b cells. Methods 2013, 64, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhao, X.; Gu, Z. Advanced optoelectronic nanodevices and nanomaterials for sensing inside single living cell. Opt. Commun. 2016. [Google Scholar] [CrossRef]
- Lal, R.; Ramachandran, S.; Arnsdorf, M.F. Multidimensional atomic force microscopy: A versatile novel technology for nanopharmacology research. AAPS J. 2010, 12, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Cho, C.H.; Park, E.K.; Jung, M.-H.; Yoon, K.-S.; Park, H.-K. Afm-detected apoptotic changes in morphology and biophysical property caused by paclitaxel in ishikawa and hela cells. PLoS ONE 2012, 7, e30066. [Google Scholar] [CrossRef] [PubMed]
- González-Solís, J.; Luévano-Colmenero, G.; Vargas-Mancilla, J. Surface enhanced raman spectroscopy in breast cancer cells. Laser Ther. 2013, 22, 37–42. [Google Scholar] [PubMed]
- Kumar, N.; Mignuzzi, S.; Su, W.; Roy, D. Tip-enhanced raman spectroscopy: Principles and applications. EPJ Tech. Instrum. 2015, 2, 1. [Google Scholar] [CrossRef]
- Zhu, Z.; Frey, O.; Haandbaek, N.; Franke, F.; Rudolf, F.; Hierlemann, A. Time-lapse electrical impedance spectroscopy for monitoring the cell cycle of single immobilized s. Pombe cells. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.; Jang, L.S.; Tsai, S.L.; Chen, M.K.; Wang, M.H. Impedance analysis of single melanoma cells in microfluidic devices. Electroanalysis 2014, 26, 2129–2137. [Google Scholar] [CrossRef]
- Giliberti, V.; Baldassarre, L.; Rosa, A.; de Turris, V.; Ortolani, M.; Calvani, P.; Nucara, A. Protein clustering in chemically stressed hela cells studied by infrared nanospectroscopy. Nanoscale 2016, 8, 17560–17567. [Google Scholar] [CrossRef] [PubMed]
- Perlaki, C.M.; Liu, Q.; Lim, M. Raman spectroscopy based techniques in tissue engineering—An overview. Appl. Spectrosc. Rev. 2014, 49, 513–532. [Google Scholar] [CrossRef]
- Li, M.; Xu, J.; Romero-Gonzalez, M.; Banwart, S.A.; Huang, W.E. Single cell raman spectroscopy for cell sorting and imaging. Curr. Opin. Biotechnol. 2012, 23, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Boardman, D.G.; Ward, A.; Huang, W.E. Single-cell raman sorting. Environ. Microbiol. Methods Protoc. 2014, 147–153. [Google Scholar]
- Wang, Y.; Song, Y.; Thompson, I.P.; Xu, J.; Huang, W.E. Single-cell metabolomics. Hydrocarb. Lipid Microbiol. Protoc. 2016, 77–90. [Google Scholar]
- Smith, E.; Dent, G. Modern Raman Spectroscopy a Practical Approach; John Wiley and Sons: West Sussex, UK, 2005. [Google Scholar]
- Huang, W.E.; Griffiths, R.I.; Thompson, I.P.; Bailey, M.J.; Whiteley, A.S. Raman microscopic analysis of single microbial cells. Anal. Chem. 2004, 76, 4452–4458. [Google Scholar] [CrossRef] [PubMed]
- Palonpon, A.F.; Sodeoka, M.; Fujita, K. Molecular imaging of live cells by raman microscopy. Curr. Opin. Chem. Biol. 2013, 17, 708–715. [Google Scholar] [CrossRef] [PubMed]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L. Using raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.; Wright, K.L.; Ashton, L. Raman spectroscopy: An evolving technique for live cell studies. Analyst 2016, 141, 3590–3600. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, M.; Hendra, P.J.; McQuillan, A. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 1974, 26, 163–166. [Google Scholar] [CrossRef]
- Jeanmaire, D.L.; Van Duyne, R.P. Surface raman spectroelectrochemistry: Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J. Electroanal. Chem. Interfacial Electrochem. 1977, 84, 1–20. [Google Scholar] [CrossRef]
- Kambhampati, P.; Child, C.; Foster, M.C.; Campion, A. On the chemical mechanism of surface enhanced raman scattering: Experiment and theory. J. Chem. Phys. 1998, 108, 5013–5026. [Google Scholar] [CrossRef]
- Schatz, G.C.; Van Duyne, R.P. Electromagnetic mechanism of surface-enhanced spectroscopy. In Handbook of Vibrational Spectroscopy; John Wiley & Sons Ltd.: Chichester, UK, 2002. [Google Scholar]
- Kneipp, K.; Wang, Y.; Kneipp, H.; Perelman, L.T.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Single molecule detection using surface-enhanced raman scattering (SERs). Phys. Rev. Lett. 1997, 78, 1667. [Google Scholar] [CrossRef]
- Kneipp, K.; Kneipp, H.; Manoharan, R.; Hanlon, E.B.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Extremely large enhancement factors in surface-enhanced raman scattering for molecules on colloidal gold clusters. Appl. Spectrosc. 1998, 52, 1493–1497. [Google Scholar] [CrossRef]
- Sharma, B.; Frontiera, R.R.; Henry, A.-I.; Ringe, E.; Van Duyne, R.P. Sers: Materials, applications, and the future. Mater. Today 2012, 15, 16–25. [Google Scholar] [CrossRef]
- Fan, M. Surface enhanced raman scattering (sers) nanoprobes as cancer theranostics. In Nanomaterials for Tumor Targeting Theranostics: A Proactive Clinical Perspective; Tan, M., Wu, A., Eds.; World Scientific Publishing: Singapore, 2016; pp. 177–204. [Google Scholar]
- Prochazka, M. Biomolecular sers applications. In Surface-Enhanced Raman Spectroscopy; Springer: Cham, Switzerland, 2016; pp. 93–125. [Google Scholar]
- Kittler, S.; Greulich, C.; Diendorf, J.; Koller, M.; Epple, M. Toxicity of silver nanoparticles increases during storage because of slow dissolution under release of silver ions. Chem. Mater. 2010, 22, 4548–4554. [Google Scholar] [CrossRef]
- Mert, S.; Çulha, M. Surface-enhanced raman scattering-based detection of cancerous renal cells. Appl. Spectrosc. 2014, 68, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Chen, R.; Feng, S.; Li, Y.; Huang, Z.; Xie, S.; Yu, Y.; Cheng, M.; Zeng, H. Rapid delivery of silver nanoparticles into living cells by electroporation for surface-enhanced raman spectroscopy. Biosens. Bioelectron. 2009, 25, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Kneipp, K.; Kneipp, H.; Kneipp, J. Surface-enhanced raman scattering in local optical fields of silver and gold nanoaggregates from single-molecule raman spectroscopy to ultrasensitive probing in live cells. Acc. Chem. Res. 2006, 39, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Stamplecoskie, K.G.; Scaiano, J.C.; Tiwari, V.S.; Anis, H. Optimal size of silver nanoparticles for surface-enhanced raman spectroscopy. J. Phys. Chem. C 2011, 115, 1403–1409. [Google Scholar] [CrossRef]
- Bell, S.E.; McCourt, M.R. Sers enhancement by aggregated au colloids: Effect of particle size. Phys. Chem. Chem. Phys. 2009, 11, 7455–7462. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-H.; Park, J.; Dembereldorj, U.; Cho, K.; Lee, K.; Yang, S.I.; Lee, S.Y.; Joo, S.-W. Raman detection of localized transferrin-coated gold nanoparticles inside a single cell. Anal. Bioanal. Chem. 2011, 401, 1631–1639. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.-Q.; Yi, J.; Hu, S.; Liu, B.-J.; Liu, J.-Y.; Wang, X.; Ren, B. Size effect on sers of gold nanorods demonstrated via single nanoparticle spectroscopy. J. Phys. Chem. C 2016, 120, 20806–20813. [Google Scholar] [CrossRef]
- Tian, F.; Bonnier, F.; Casey, A.; Shanahan, A.E.; Byrne, H.J. Surface enhanced raman scattering with gold nanoparticles: Effect of particle shape. Anal. Methods 2014, 6, 9116–9123. [Google Scholar] [CrossRef]
- Xie, J.; Zhang, Q.; Lee, J.Y.; Wang, D.I. The synthesis of sers-active gold nanoflower tags for in vivo applications. ACS Nano 2008, 2, 2473–2480. [Google Scholar] [CrossRef] [PubMed]
- Fales, A.M.; Yuan, H.; Vo-Dinh, T. Development of hybrid silver-coated gold nanostars for nonaggregated surface-enhanced raman scattering. J. Phys. Chem. C 2014, 118, 3708–3715. [Google Scholar] [CrossRef] [PubMed]
- Mahyari, F.A.; Tohidi, M.; Safavi, A. Synthesis of gold nanoflowers using deep eutectic solvent with high surface enhanced raman scattering properties. Mater. Res. Express 2016, 3, 095006. [Google Scholar] [CrossRef]
- Giannini, V.; Rodríguez-Oliveros, R.; Sánchez-Gil, J.A. Surface plasmon resonances of metallic nanostars/nanoflowers for surface-enhanced raman scattering. Plasmonics 2010, 5, 99–104. [Google Scholar] [CrossRef]
- Mohan, S.; Subramanian, B. Surface enhanced raman scattering studies of silver-gold normal and inverted core-shell nanostructures on their efficiency of detecting molecules. Procedia Eng. 2014, 92, 19–25. [Google Scholar] [CrossRef]
- Wang, S.-H.; Lee, C.-W.; Chiou, A.; Wei, P.-K. Size-dependent endocytosis of gold nanoparticles studied by three-dimensional mapping of plasmonic scattering images. J. Nanobiotechnol. 2010, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wu, Y.; Jin, S.; Tian, Y.; Zhang, X.; Zhao, Y.; Yu, L.; Liang, X.-J. Gold nanoparticles induce autophagosome accumulation through size-dependent nanoparticle uptake and lysosome impairment. ACS Nano 2011, 5, 8629–8639. [Google Scholar] [CrossRef] [PubMed]
- Sadauskas, E.; Wallin, H.; Stoltenberg, M.; Vogel, U.; Doering, P.; Larsen, A.; Danscher, G. Kupffer cells are central in the removal of nanoparticles from the organism. Part. Fibre Toxicol. 2007, 4. [Google Scholar] [CrossRef] [PubMed]
- Dykman, L.A.; Khlebtsov, N.G. Uptake of engineered gold nanoparticles into mammalian cells. Chem. Rev. 2013, 114, 1258–1288. [Google Scholar] [CrossRef] [PubMed]
- Nabiev, I.; Morjani, H.; Manfait, M. Selective analysis of antitumor drug interaction with living cancer cells as probed by surface-enhanced raman spectroscopy. Eur. Biophys. J. 1991, 19, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Kneipp, K.; Haka, A.S.; Kneipp, H.; Badizadegan, K.; Yoshizawa, N.; Boone, C.; Shafer-Peltier, K.E.; Motz, J.T.; Dasari, R.R.; Feld, M.S. Surface-enhanced raman spectroscopy in single living cells using gold nanoparticles. Appl. Spectrosc. 2002, 56, 150–154. [Google Scholar] [CrossRef]
- Talley, C.E.; Jusinski, L.; Hollars, C.W.; Lane, S.M.; Huser, T. Intracellular ph sensors based on surface-enhanced raman scattering. Anal. Chem. 2004, 76, 7064–7068. [Google Scholar] [CrossRef] [PubMed]
- Eliasson, C.; Lorén, A.; Engelbrektsson, J.; Josefson, M.; Abrahamsson, J.; Abrahamsson, K. Surface-enhanced raman scattering imaging of single living lymphocytes with multivariate evaluation. Spectrochim. Acta Part A 2005, 61, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; El-Sayed, I.H.; Qian, W.; El-Sayed, M.A. Cancer cells assemble and align gold nanorods conjugated to antibodies to produce highly enhanced, sharp, and polarized surface raman spectra: A potential cancer diagnostic marker. Nano Lett. 2007, 7, 1591–1597. [Google Scholar] [CrossRef] [PubMed]
- Shamsaie, A.; Jonczyk, M.; Sturgis, J.; Robinson, J.P.; Irudayaraj, J. Intracellularly grown gold nanoparticles as potential surface-enhanced raman scattering probes. J. Biomed. Opt. 2007, 12. [Google Scholar] [CrossRef] [PubMed]
- Shamsaie, A.; Heim, J.; Yanik, A.A.; Irudayaraj, J. Intracellular quantification by surface enhanced raman spectroscopy. Chem. Phys. Lett. 2008, 461, 131–135. [Google Scholar] [CrossRef]
- Fujita, K.; Ishitobi, S.; Hamada, K.; Smith, N.I.; Taguchi, A.; Inouye, Y.; Kawata, S. Time-resolved observation of surface-enhanced raman scattering from gold nanoparticles during transport through a living cell. J. Biomed. Opt. 2009, 14. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Huefner, A.; Li, L.; Wingfield, J.; Mahajan, S. Nanoparticles and intracellular applications of surface-enhanced raman spectroscopy. Analyst 2016, 141, 5037–5055. [Google Scholar] [CrossRef] [PubMed]
- Kuku, G.; Saricam, M.; Akhatova, F.; Danilushkina, A.; Fakhrullin, R.F.; Culha, M. Surface-enhanced raman scattering to evaluate nanomaterial cytotoxicity on living cells. Anal. Chem. 2016, 88, 9813–9820. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Kuang, H.; Xu, C.; Ma, W.; Wang, L.; Kotov, N.A. Regiospecific plasmonic assemblies for in situ raman spectroscopy in live cells. J. Am. Chem. Soc. 2012, 134, 1699–1709. [Google Scholar] [CrossRef] [PubMed]
- Jaworska, A.; Jamieson, L.E.; Malek, K.; Campbell, C.J.; Choo, J.; Chlopicki, S.; Baranska, M. Sers-based monitoring of the intracellular ph in endothelial cells: The influence of the extracellular environment and tumour necrosis factor-α. Analyst 2015, 140, 2321–2329. [Google Scholar] [CrossRef] [PubMed]
- Sakhrani, N.M.; Padh, H. Organelle targeting: Third level of drug targeting. Drug Des. Dev. Ther. 2013, 7, 585–599. [Google Scholar]
- Yousif, L.F.; Stewart, K.M.; Kelley, S.O. Targeting mitochondria with organelle-specific compounds: Strategies and applications. ChemBioChem 2009, 10, 1939–1950. [Google Scholar] [CrossRef] [PubMed]
- Macara, I.G. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001, 65, 570–594. [Google Scholar] [CrossRef] [PubMed]
- Oyelere, A.K.; Chen, P.C.; Huang, X.; El-Sayed, I.H.; El-Sayed, M.A. Peptide-conjugated gold nanorods for nuclear targeting. Bioconj. Chem. 2007, 18, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.; Mackey, M.A.; El-Sayed, M.A. Nuclear targeting of gold nanoparticles in cancer cells induces DNA damage, causing cytokinesis arrest and apoptosis. J. Am. Chem. Soc. 2010, 132, 1517–1519. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.; Austin, L.A.; El-Sayed, M.A. Observing real-time molecular event dynamics of apoptosis in living cancer cells using nuclear-targeted plasmonically enhanced raman nanoprobes. ACS Nano 2014, 8, 4883–4892. [Google Scholar] [CrossRef] [PubMed]
- Huefner, A.; Kuan, W.-L.; Barker, R.A.; Mahajan, S. Intracellular sers nanoprobes for distinction of different neuronal cell types. Nano Lett. 2013, 13, 2463–2470. [Google Scholar] [CrossRef] [PubMed]
- Austin, L.A.; Kang, B.; El-Sayed, M.A. A new nanotechnology technique for determining drug efficacy using targeted plasmonically enhanced single cell imaging spectroscopy. J. Am. Chem. Soc. 2013, 135, 4688–4691. [Google Scholar] [CrossRef] [PubMed]
- Aioub, M.; Kang, B.; Mackey, M.A.; El-Sayed, M.A. Biological targeting of plasmonic nanoparticles improves cellular imaging via the enhanced scattering in the aggregates formed. J. Phys. Chem. Lett. 2014, 5, 2555–2561. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, J.; Lin, J.; Lin, D.; Chen, W.; Feng, S.; Huang, Z.; Li, Y.; Huang, H.; Shi, H. An optimized electroporation method for delivering nanoparticles into living cells for surface-enhanced raman scattering imaging. Appl. Phys. Lett. 2016, 108, 153701. [Google Scholar] [CrossRef]

| AuNPs Size/Shape | Surface Chemistry | Cell Line | Uptake Efficiency | Ref. |
|---|---|---|---|---|
| 15, 50 and 100 nm spherical | Tri sodium Citrate | Intestinal epithelial (Caco-2) cells | Uptake and spread of 15 nm size was more rapid. Best uptake efficiency was observed with 50 nm size whereas 100 nm size tended to accumulate. | [39] |
| 10 and 25 nm spherical | Bovine Serum Albumin | Human cervical (HeLa) cancer cells | 10 nm was taken up more rapidly than 25 nm. 25 nm accumulated in higher concentrations had better retention. | [40] |
| 10, 25, and 50 nm spherical | 10-Hydroxy camptothecin (HCPT) | Human breast cancer (MDA-MB-23) cells | The 50 nm HCPT-loaded AuNPs had unique advantages over smaller NPs in terms of killing MDA-MB-231 cells due to the higher uptake efficiency | [41] |
| 2, 4, and 6 nm spherical | Cationic, neutral and anionic | Human cervical (HeLa) cells | The increasing particle size resulted in increased uptake for cationic nanoparticles whereas for neutral and anionic particles it decreased uptake efficiency. | [42] |
| 15, 40 and 80 nm spherical | Tri sodium Citrate | N9 phagocytic microglial cells, Nonphagocytic neural SH cells | SH cells engulfed small particles between 15 and 40 nm in diameter while the larger particles with a diameter of 80 nm preferentially internalized into N9 phagocytic microglial cells. | [43] |
| 7 and 14 nm diameter Rod shape | CTAB, oleate or BSA | Hematopoietic stem cell (HSC) and Human liver cancer (HepG2) cells | 7 nm gold nanorods showed higher cell uptake compared to 14 nm independent of the surface modification. | [45] |
| 40 nm diameter Rod shape | Silica coated Silica coated-folic acid modified | Human liver cancer (HepG2) cells | The cellular uptake of AuNRs@SiO2-FA was rapid while unmodified AuNRs@SiO2 showed no obvious binding or internalization. | [46] |
| Nanocages and Nanorods. | PEG coated | Human umbilical vein endothelial (HUVEC) and DU145 prostate cancer cells | Both nanocages and nanorods were taken up by HUVEC more than DU145 cells. The internalization of nanocages into DU145 cells was higher than nanorods. | [47] |
| 34 nm Nano stars and 61 nm Nanospheres | HEPES (Nanostar) and Citrate (Nanospheres) | Fibroblast cells and microvascular endothelial (RFPECs) cells | Nanospheres have higher toxicity compared to nanostars. | [37] |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altunbek, M.; Kuku, G.; Culha, M. Gold Nanoparticles in Single-Cell Analysis for Surface Enhanced Raman Scattering. Molecules 2016, 21, 1617. https://doi.org/10.3390/molecules21121617
Altunbek M, Kuku G, Culha M. Gold Nanoparticles in Single-Cell Analysis for Surface Enhanced Raman Scattering. Molecules. 2016; 21(12):1617. https://doi.org/10.3390/molecules21121617
Chicago/Turabian StyleAltunbek, Mine, Gamze Kuku, and Mustafa Culha. 2016. "Gold Nanoparticles in Single-Cell Analysis for Surface Enhanced Raman Scattering" Molecules 21, no. 12: 1617. https://doi.org/10.3390/molecules21121617
APA StyleAltunbek, M., Kuku, G., & Culha, M. (2016). Gold Nanoparticles in Single-Cell Analysis for Surface Enhanced Raman Scattering. Molecules, 21(12), 1617. https://doi.org/10.3390/molecules21121617






