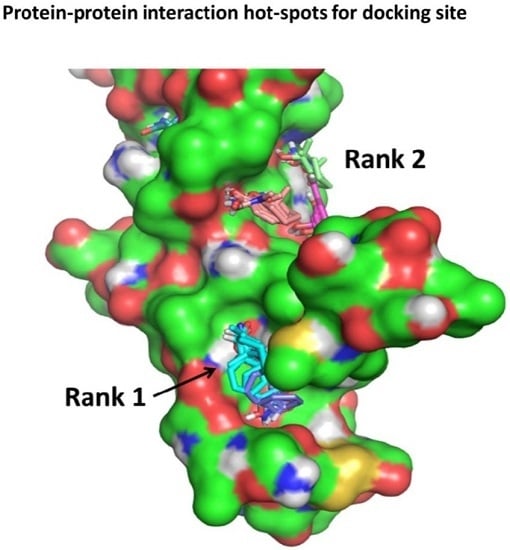
Surfing the Protein-Protein Interaction Surface Using Docking Methods: Application to the Design of PPI Inhibitors
Abstract
:1. Introduction
2. Protein-Protein Interactions
3. Identification of Hot Spots on Proteins and Docking in PPI
| Method | Website | Type | Reference |
|---|---|---|---|
| FunHunt | http://funhunt.furmanlab.cs.huji.ac.il/ | Online | [128] |
| Fiberdock | http://bioinfo3d.cs.tau.ac.il/FiberDock/ | Online | [129,130] |
| TACOS | http://zhanglab.ccmb.med.umich.edu/TACOS/about.html | Online | [131] |
| SPA-PP | https://www.dl.dropboxusercontent.com/u/1865642/Optimization.cpp. | Software | [132] |
| FastContact | http://structure.pitt.edu/servers/fastcontact/ | Online | [133] |
| CONSRANK | www.molnac.unisa.it/BioTools/consrank/ | Online | [134] |
| FILTREST 3D | www.genesilico.pl/software/stand-alone/filtrest3d | Software | [135] |
3.1. Identification of Hot Spots and Druggability
| Method | Website | Type | Reference |
|---|---|---|---|
| FTMap | http://ftmap.bu.edu/login.php | webserver | [150] |
| PredHS | http://www.predhs.org/ | webserver | [154] |
| HSPred | http://bioinf.cs.ucl.ac.uk/structure/ | webserver | [155] |
| iPred | http://modlab-cadd.ethz.ch/software/ipred/ | Java based webtool | [156] |
| HotPoint | http://prism.ccbb.ku.edu.tr/hotpoint/ | webserver | [157] |
| ROBETTA | http://robetta.bakerlab.org/ | webserver | [158] |
| KFC2 | http://kfc.mitchell-lab.org/ | webserver | [159] |
| PCRPi | http://www.bioinsilico.org/PCRPi/ | webserver | [160] |
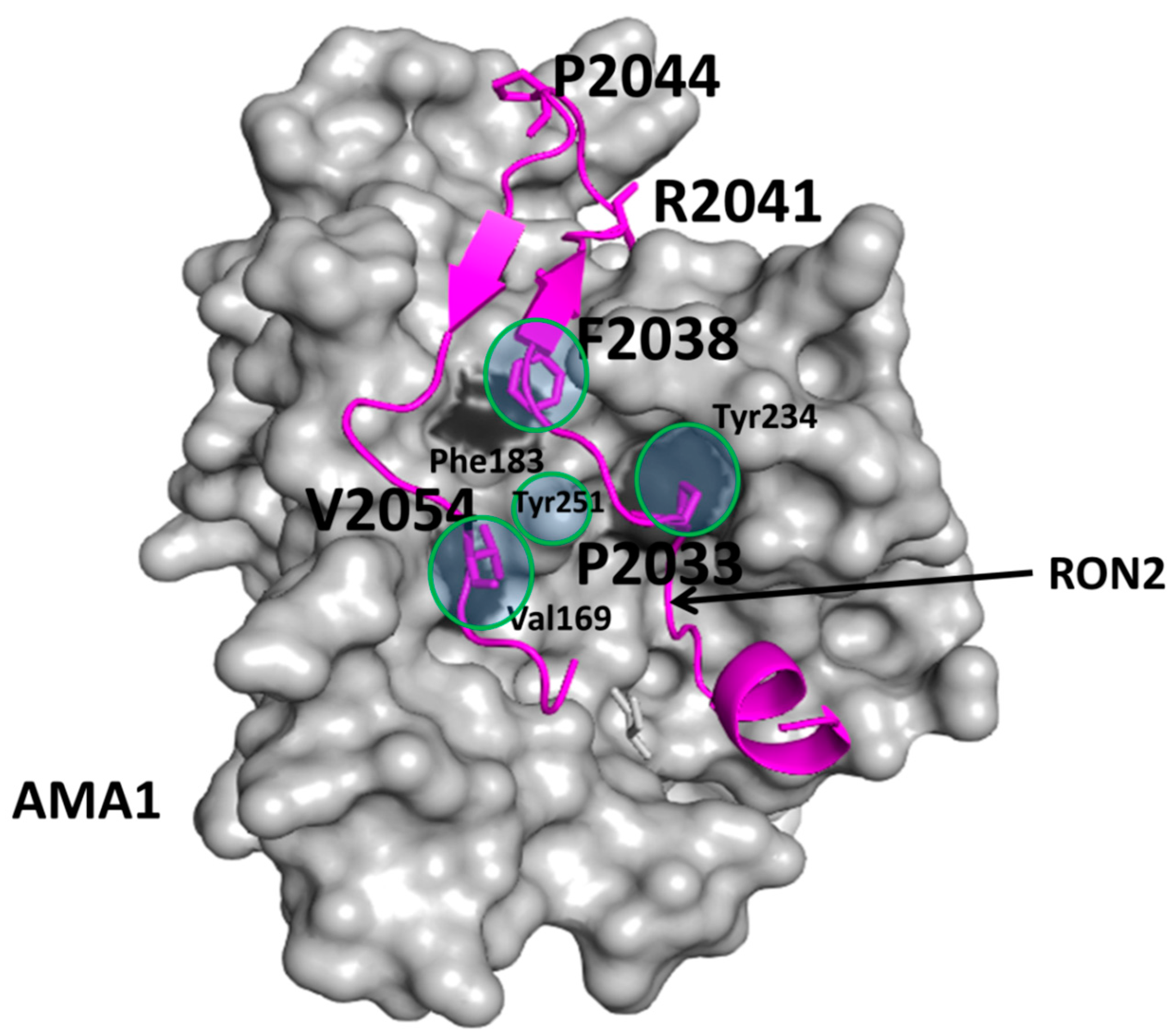
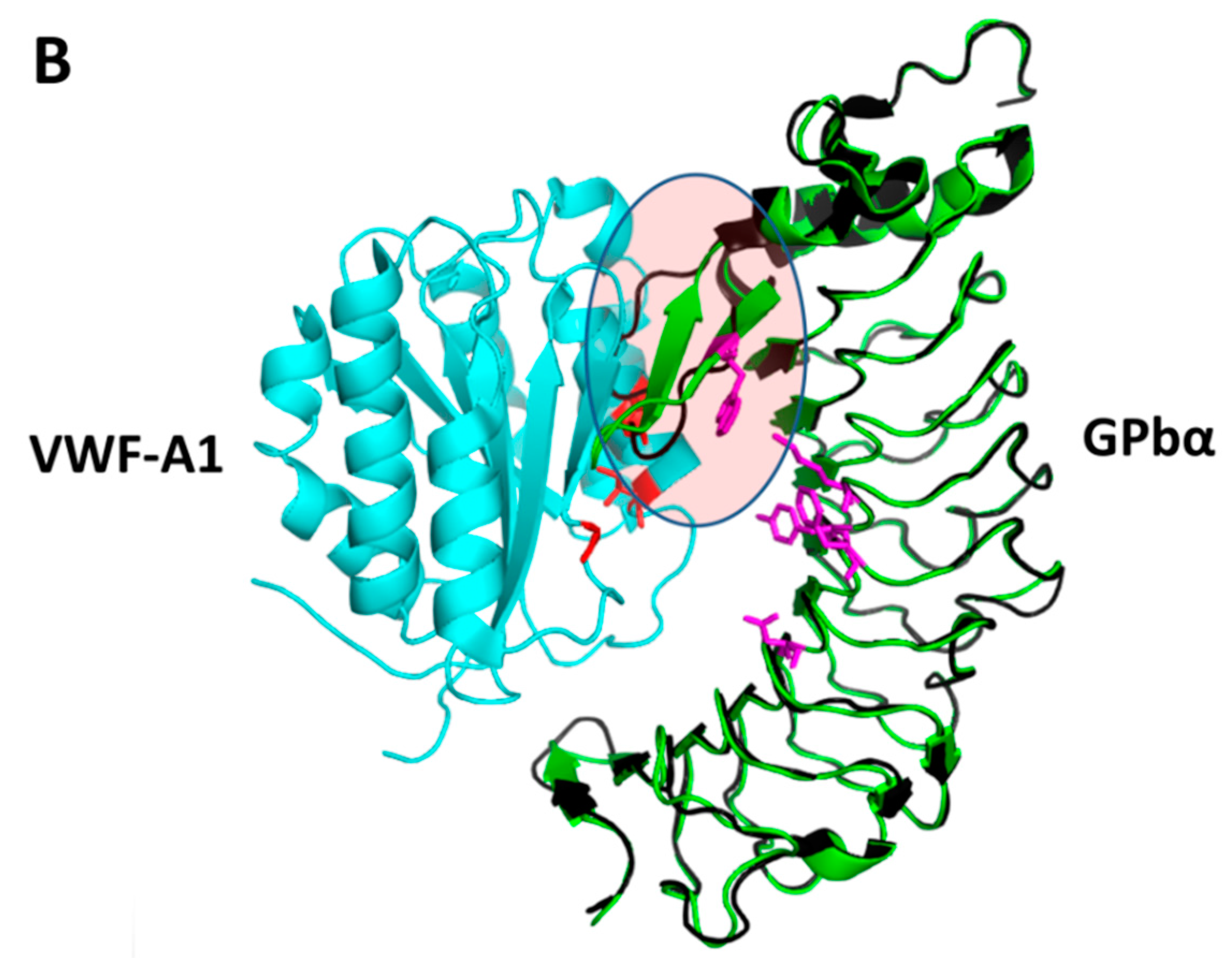
3.2. Examples of Design of PPI Inhibitors

4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Kuntz, I.D.; Blaney, J.M.; Oatley, S.J.; Langridge, R.; Ferrin, T.E. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982, 161, 269–288. [Google Scholar] [CrossRef]
- Venhorst, J.; ter Laak, A.M.; Commandeur, J.N.; Funae, Y.; Hiroi, T.; Vermeulen, N.P. Homology modeling of rat and human cytochrome p450 2d (cyp2d) isoforms and computational rationalization of experimental ligand-binding specificities. J. Med. Chem. 2003, 46, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayanajois, S.D.; Hill, R.A. Medicinal chemistry for 2020. Future Med. Chem. 2011, 3, 1765–1786. [Google Scholar] [CrossRef] [PubMed]
- Fotouhi, N.; Gillespie, P.; Goodnow, R., Jr. Lead generation: Reality check on commonly held views. Expert Opin. Drug Discov. 2008, 3, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Light, D.W.; Warburton, R. Demythologizing the high costs of pharmaceutical research. BioSocieties 2011, 6, 34–50. [Google Scholar] [CrossRef]
- Schneider, G. Virtual screening: An endless staircase? Nat. Rev. Drug Discov. 2010, 9, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.H.; Ma, D.L. Recent advances in virtual screening for drug discovery. Methods 2015, 71, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G.; Fechner, U. Computer-based de novo design of drug-like molecules. Nat. Rev. Drug Discov. 2005, 4, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Leach, A.R.; Shoichet, B.K.; Peishoff, C.E. Prediction of protein-ligand interactions. Docking and scoring: Successes and gaps. J. Med. Chem. 2006, 49, 5851–5855. [Google Scholar] [CrossRef] [PubMed]
- Moitessier, N.; Englebienne, P.; Lee, D.; Lawandi, J.; Corbeil, C.R. Towards the development of universal, fast and highly accurate docking/scoring methods: A long way to go. Br. J. Pharmacol. 2008, 153 (Suppl. 1), S7–S26. [Google Scholar] [CrossRef] [PubMed]
- Grinter, S.Z.; Zou, X. Challenges, applications, and recent advances of protein-ligand docking in structure-based drug design. Molecules 2014, 19, 10150–10176. [Google Scholar] [CrossRef] [PubMed]
- Perola, E.; Walters, W.P.; Charifson, P.S. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins 2004, 56, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Thiel, P.; Kaiser, M.; Ottmann, C. Small-molecule stabilization of protein-protein interactions: An underestimated concept in drug discovery? Angew. Chem. 2012, 51, 2012–2018. [Google Scholar] [CrossRef] [PubMed]
- Stumpf, M.P.; Thorne, T.; de Silva, E.; Stewart, R.; An, H.J.; Lappe, M.; Wiuf, C. Estimating the size of the human interactome. Proc. Natl. Acad. Sci. USA 2008, 105, 6959–6964. [Google Scholar] [CrossRef] [PubMed]
- Berg, T. Small-molecule inhibitors of protein-protein interactions. Curr. Opin. Drug Discov. Dev. 2008, 11, 666–674. [Google Scholar]
- Fry, D.C. Drug-like inhibitors of protein-protein interactions: A structural examination of effective protein mimicry. Curr. Protein Pept. Sci. 2008, 9, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Fuller, J.C.; Burgoyne, N.J.; Jackson, R.M. Predicting druggable binding sites at the protein-protein interface. Drug Discov. Today 2009, 14, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Sawyer, N.; Regan, L. Protein-protein interactions: General trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 2013, 22, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Reichmann, D.; Rahat, O.; Cohen, M.; Neuvirth, H.; Schreiber, G. The molecular architecture of protein-protein binding sites. Curr. Opin. Struct. Biol. 2007, 17, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Moreira, I.S.; Fernandes, P.A.; Ramos, M.J. Hot spots—A review of the protein-protein interface determinant amino-acid residues. Proteins 2007, 68, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Lo Conte, L.; Chothia, C.; Janin, J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999, 285, 2177–2198. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.C.; Coleman, R.G.; Smyth, K.T.; Cao, Q.; Soulard, P.; Caffrey, D.R.; Salzberg, A.C.; Huang, E.S. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 2007, 25, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L.; Groom, C.R. The druggable genome. Nat. Rev. Drug Discov. 2002, 1, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.D.; Hu, L.; Falkner, J.A.; Benson, M.L.; Nerothin, J.P.; Carlson, H.A. Exploring protein-ligand recognition with binding moad. J. Mol. Graph. Model. 2006, 24, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.A.; McClendon, C.L. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 2007, 450, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. Drugbank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008, 36, D901–D906. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.G.; Guo, J.; Wu, Y. Chemokine receptor ccr5 antagonist maraviroc: Medicinal chemistry and clinical applications. Curr. Top. Med. Chem. 2014, 14, 1504–1514. [Google Scholar] [CrossRef] [PubMed]
- Domling, A. Small molecular weight protein-protein interaction antagonists: An insurmountable challenge? Curr. Opin. Chem. Biol. 2008, 12, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Clackson, T.; Wells, J.A. A hot spot of binding energy in a hormone-receptor interface. Science 1995, 267, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, P.; Janin, J. Dissecting protein-protein recognition sites. Proteins 2002, 47, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Chene, P. Drugs targeting protein-protein interactions. ChemMedChem 2006, 1, 400–411. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Unraveling hot spots in binding interfaces: Progress and challenges. Curr. Opin. Struct. Biol. 2002, 12, 14–20. [Google Scholar] [CrossRef]
- Bogan, A.A.; Thorn, K.S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998, 280, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Meireles, L.M.; Mustata, G. Discovery of modulators of protein-protein interactions: Current approaches and limitations. Curr. Top. Med. Chem. 2011, 11, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Buchwald, P. Small-molecule protein-protein interaction inhibitors: Therapeutic potential in light of molecular size, chemical space, and ligand binding efficiency considerations. IUBMB Life 2010, 62, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Ruiz, D.; Gohlke, H. Targeting protein-protein interactions with small molecules: Challenges and perspectives for computational binding epitope detection and ligand finding. Curr. Med. Chem. 2006, 13, 2607–2625. [Google Scholar] [CrossRef] [PubMed]
- Sperandio, O.; Reynes, C.H.; Camproux, A.C.; Villoutreix, B.O. Rationalizing the chemical space of protein-protein interaction inhibitors. Drug Discov. Today 2010, 15, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Nero, T.L.; Morton, C.J.; Holien, J.K.; Wielens, J.; Parker, M.W. Oncogenic protein interfaces: Small molecules, big challenges. Nat. Rev. Cancer 2014, 14, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J. Inhibition of protein-protein interactions using designed molecules. Chem. Soc. Rev. 2009, 38, 3289–3300. [Google Scholar] [CrossRef] [PubMed]
- Fischer, G.; Rossmann, M.; Hyvonen, M. Alternative modulation of protein-protein interactions by small molecules. Curr. Opin. Biotechnol. 2015, 35, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.M.; Van Molle, I.; Baud, M.G.; Galdeano, C.; Geraldes, C.F.; Ciulli, A. Is nmr fragment screening fine-tuned to assess druggability of protein-protein interactions? ACS Med. Chem. Lett. 2014, 5, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Kuenemann, M.A.; Sperandio, O.; Labbe, C.M.; Lagorce, D.; Miteva, M.A.; Villoutreix, B.O. In silico design of low molecular weight protein-protein interaction inhibitors: Overall concept and recent advances. Prog. Biophys. Mol. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Jubb, H.; Blundell, T.L.; Ascher, D.B. Flexibility and small pockets at protein-protein interfaces: New insights into druggability. Prog. Biophys. Mol. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Aguilar, A.; Bernard, D.; Wang, S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 inhibitors) in clinical trials for cancer treatment. J. Med. Chem. 2015, 58, 1038–1052. [Google Scholar] [CrossRef] [PubMed]
- Cierpicki, T.; Grembecka, J. Challenges and opportunities in targeting the menin-mll interaction. Future Med. Chem. 2014, 6, 447–462. [Google Scholar] [CrossRef] [PubMed]
- Falchi, F.; Caporuscio, F.; Recanatini, M. Structure-based design of small-molecule protein-protein interaction modulators: The story so far. Future Med. Chem. 2014, 6, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Vignaroli, G.; Botta, M. Small molecules modulation of 14–3-3 protein-protein interactions. Drug Discov. Today Technol. 2013, 10, e541–e547. [Google Scholar] [CrossRef] [PubMed]
- Silvian, L.; Enyedy, I.; Kumaravel, G. Inhibitors of protein-protein interactions: New methodologies to tackle this challenge. Drug Discov. Today Technol. 2013, 10, e509–e515. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Wang, W.; Fang, G. Targeting protein-protein interaction by small molecules. Annu. Rev. Pharmacol. Toxicol. 2014, 54, 435–456. [Google Scholar] [CrossRef] [PubMed]
- Coelho, E.D.; Arrais, J.P.; Oliveira, J.L. From protein-protein interactions to rational drug design: Are computational methods up to the challenge? Curr. Top. Med. Chem. 2013, 13, 602–618. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.; Phillips, A.T.; Weng, Z. Structure prediction of protein complexes. In Computational Methods for Protein Structure Prediction and Modeling; Xu, Y., Xu, D., Liang, J., Eds.; Springer: New York, NY, USA, 2007; Volume 2, pp. 109–134. [Google Scholar]
- Krogan, N.J.; Cagney, G.; Yu, H.; Zhong, G.; Guo, X.; Ignatchenko, A.; Li, J.; Pu, S.; Datta, N.; Tikuisis, A.P.; et al. Global landscape of protein complexes in the yeast saccharomyces cerevisiae. Nature 2006, 440, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.; Ideker, T.; Karp, R.M.; Sharan, R. Efficient algorithms for detecting signaling pathways in protein interaction networks. J. Comput. Biol. 2006, 13, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Sharan, R.; Ulitsky, I.; Shamir, R. Network-based prediction of protein function. Mol. Syst. Biol. 2007, 3, 88. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Cusick, M.E.; Barabasi, A.L. Interactome networks and human disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Wodak, S.J.; Vlasblom, J.; Turinsky, A.L.; Pu, S. Protein-protein interaction networks: The puzzling riches. Curr. Opin. Struct. Biol. 2013, 23, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Basse, M.J.; Betzi, S.; Bourgeas, R.; Bouzidi, S.; Chetrit, B.; Hamon, V.; Morelli, X.; Roche, P. 2p2idb: A structural database dedicated to orthosteric modulation of protein-protein interactions. Nucleic Acids Res. 2013, 41, D824–D827. [Google Scholar] [CrossRef] [PubMed]
- Morelli, X.; Bourgeas, R.; Roche, P. Chemical and structural lessons from recent successes in protein-protein interaction inhibition (2p2i). Curr. Opin. Chem. Biol. 2011, 15, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.C.; Petrey, D.; Deng, L.; Qiang, L.; Shi, Y.; Thu, C.A.; Bisikirska, B.; Lefebvre, C.; Accili, D.; Hunter, T.; et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature 2012, 490, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. String v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The string database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, B.A.; Zhang, D.; Tyagi, M.; Thangudu, R.R.; Fong, J.H.; Marchler-Bauer, A.; Bryant, S.H.; Madej, T.; Panchenko, A.R. Ibis (inferred biomolecular interaction server) reports, predicts and integrates multiple types of conserved interactions for proteins. Nucleic Acids Res. 2012, 40, D834–D840. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, B.A.; Zhang, D.; Thangudu, R.R.; Tyagi, M.; Fong, J.H.; Marchler-Bauer, A.; Bryant, S.H.; Madej, T.; Panchenko, A.R. Inferred biomolecular interaction server—A web server to analyze and predict protein interacting partners and binding sites. Nucleic Acids Res. 2010, 38, D518–D524. [Google Scholar] [CrossRef] [PubMed]
- McDowall, M.D.; Scott, M.S.; Barton, G.J. Pips: Human protein-protein interaction prediction database. Nucleic Acids Res. 2009, 37, D651–D656. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.S.; Barton, G.J. Probabilistic prediction and ranking of human protein-protein interactions. BMC Bioinform. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.C.; Petrey, D.; Norel, R.; Honig, B.H. Protein interface conservation across structure space. Proc. Natl. Acad. Sci. USA 2010, 107, 10896–10901. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Zhang, Q.C.; Dey, F.; Chen, B.Y.; Honig, B.; Petrey, D. Markus: A server to navigate sequence-structure-function space. Nucleic Acids Res. 2011, 39, W357–W361. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.J.; Xenarios, I.; Eisenberg, D. Describing biological protein interactions in terms of protein states and state transitions: The livedip database. Mol. Cell. Proteomics MCP 2002, 1, 104–116. [Google Scholar] [CrossRef]
- Bader, G.D.; Betel, D.; Hogue, C.W. Bind: The biomolecular interaction network database. Nucleic Acids Res. 2003, 31, 248–250. [Google Scholar] [CrossRef] [PubMed]
- Guldener, U.; Munsterkotter, M.; Oesterheld, M.; Pagel, P.; Ruepp, A.; Mewes, H.W.; Stumpflen, V. Mpact: The mips protein interaction resource on yeast. Nucleic Acids Res. 2006, 34, D436–D441. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.C.; Crawford, M.E.; Hirschman, J.E.; Kranz, J.E.; Olsen, P.; Robertson, L.S.; Skrzypek, M.S.; Braun, B.R.; Hopkins, K.L.; Kondu, P.; et al. Ypd, pombepd and wormpd: Model organism volumes of the bioknowledge library, an integrated resource for protein information. Nucleic Acids Res. 2001, 29, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.C.; Hogan, J.D.; Cusick, M.E.; Davis, B.P.; Fancher, A.M.; Hodges, P.E.; Kondu, P.; Lengieza, C.; Lew-Smith, J.E.; Lingner, C.; et al. The yeast proteome database (YPD) and caenorhabditis elegans proteome database (WormPD): Comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000, 28, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Cesareni, G.; Chatr-aryamontri, A.; Licata, L.; Ceol, A. Searching the mint database for protein interaction information. Curr. Protoc. Bioinform. 2008. [Google Scholar] [CrossRef]
- Zanzoni, A.; Montecchi-Palazzi, L.; Quondam, M.; Ausiello, G.; Helmer-Citterich, M.; Cesareni, G. Mint: A molecular interaction database. FEBS Lett. 2002, 513, 135–140. [Google Scholar] [CrossRef]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The mintact project—Intact as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef] [PubMed]
- Chatr-Aryamontri, A.; Breitkreutz, B.J.; Oughtred, R.; Boucher, L.; Heinicke, S.; Chen, D.; Stark, C.; Breitkreutz, A.; Kolas, N.; O’Donnell, L.; et al. The biogrid interaction database: 2015 update. Nucleic Acids Res. 2015, 43, D470–D478. [Google Scholar] [CrossRef] [PubMed]
- Keshava Prasad, T.S.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009, 37, D767–D772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, Y.; He, F. An overview of human protein databases and their application to functional proteomics in health and disease. Sci. China Life Sci. 2011, 54, 988–998. [Google Scholar] [CrossRef] [PubMed]
- Kundrotas, P.J.; Alexov, E. Protcom: Searchable database of protein complexes enhanced with domain-domain structures. Nucleic Acids Res. 2007, 35, D575–D579. [Google Scholar] [CrossRef] [PubMed]
- Talley, K.; Ng, C.; Shoppell, M.; Kundrotas, P.; Alexov, E. On the electrostatic component of protein-protein binding free energy. PMC Biophys. 2008, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Mosca, R.; Ceol, A.; Stein, A.; Olivella, R.; Aloy, P. 3did: A catalog of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 2014, 42, D374–D379. [Google Scholar] [CrossRef] [PubMed]
- Stein, A.; Ceol, A.; Aloy, P. 3did: Identification and classification of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 2011, 39, D718–D723. [Google Scholar] [CrossRef] [PubMed]
- Davis, F.P.; Sali, A. Pibase: A comprehensive database of structurally defined protein interfaces. Bioinformatics 2005, 21, 1901–1907. [Google Scholar] [CrossRef] [PubMed]
- Forster, M.; Forster, P.; Elsharawy, A.; Hemmrich, G.; Kreck, B.; Wittig, M.; Thomsen, I.; Stade, B.; Barann, M.; Ellinghaus, D.; et al. From next-generation sequencing alignments to accurate comparison and validation of single-nucleotide variants: The pibase software. Nucleic Acids Res. 2013, 41, e16. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [PubMed]
- Hashimoto, H. Recent structural studies of carbohydrate-binding modules. Cell. Mol. Life Sci. 2006, 63, 2954–2967. [Google Scholar] [CrossRef] [PubMed]
- Winter, C.; Henschel, A.; Kim, W.K.; Schroeder, M. SCOPPI: A structural classification of protein-protein interfaces. Nucleic Acids Res. 2006, 34, D310–D314. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.K.; Ison, J.C. Survey of the geometric association of domain-domain interfaces. Proteins 2005, 61, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Miller, B.L.; Clements, J.; Bateman, A. Ipfam: A database of protein family and domain interactions found in the protein data bank. Nucleic Acids Res. 2014, 42, D364–D373. [Google Scholar] [CrossRef] [PubMed]
- Sumathy, R.; Rao, A.S.; Chandrakanth, N.; Gopalakrishnan, V.K. In silico identification of protein-protein interactions in silkworm, bombyx mori. Bioinformation 2014, 10, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.K.; Zhang, Z.; Tan, S.H.; Lin, K. Interdom: A database of putative interacting protein domains for validating predicted protein interactions and complexes. Nucleic Acids Res. 2003, 31, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Pagel, P.; Vilne, B.; Frishman, D. Dima 3.0: Domain interaction map. Nucleic Acids Res. 2011, 39, D724–D729. [Google Scholar] [CrossRef] [PubMed]
- Bowers, P.M.; Pellegrini, M.; Thompson, M.J.; Fierro, J.; Yeates, T.O.; Eisenberg, D. Prolinks: A database of protein functional linkages derived from coevolution. Genome Biol. 2004, 5, R35. [Google Scholar] [CrossRef] [PubMed]
- Kastritis, P.L.; Rodrigues, J.P.; Bonvin, A.M. Haddock(2p2i): A biophysical model for predicting the binding affinity of protein-protein interaction inhibitors. J. Chem. Inf. Model. 2014, 54, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Garcia, B.; Pons, C.; Fernandez-Recio, J. Pydockweb: A web server for rigid-body protein-protein docking using electrostatics and desolvation scoring. Bioinformatics 2013, 29, 1698–1699. [Google Scholar] [CrossRef] [PubMed]
- Pons, C.; Jimenez-Gonzalez, D.; Gonzalez-Alvarez, C.; Servat, H.; Cabrera-Benitez, D.; Aguilar, X.; Fernandez-Recio, J. Cell-dock: High-performance protein-protein docking. Bioinformatics 2012, 28, 2394–2396. [Google Scholar] [CrossRef] [PubMed]
- Ghoorah, A.W.; Devignes, M.D.; Smail-Tabbone, M.; Ritchie, D.W. Kbdock 2013: A spatial classification of 3d protein domain family interactions. Nucleic Acids Res. 2014, 42, D389–D395. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, C.; Chowdhury, R.; Siddavanahalli, V. F2dock: Fast fourier protein-protein docking. IEEEACM Trans. Comput. Biol. Bioinform. 2011, 8, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Palma, P.N.; Krippahl, L.; Wampler, J.E.; Moura, J.J. Bigger: A new (soft) docking algorithm for predicting protein interactions. Proteins 2000, 39, 372–384. [Google Scholar] [CrossRef]
- Garzon, J.I.; Lopez-Blanco, J.R.; Pons, C.; Kovacs, J.; Abagyan, R.; Fernandez-Recio, J.; Chacon, P. Frodock: A new approach for fast rotational protein-protein docking. Bioinformatics 2009, 25, 2544–2551. [Google Scholar] [CrossRef] [PubMed]
- Viswanath, S.; Ravikant, D.V.; Elber, R. Dock/pierr: Web server for structure prediction of protein-protein complexes. Methods Mol. Biol. 2014, 1137, 199–207. [Google Scholar] [PubMed]
- Pierce, B.G.; Hourai, Y.; Weng, Z. Accelerating protein docking in zdock using an advanced 3d convolution library. PLoS ONE 2011, 6, e24657. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.H.; Vreven, T.; Weng, Z. Zdock server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman, V.; Yang, Y.D.; Sael, L.; Kihara, D. Protein-protein docking using region-based 3D Zernike descriptors. BMC Bioinform. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.; Saladin, A.; Fiorucci, S.; Prevost, C.; Zacharias, M. Attract and ptools: Open source programs for protein-protein docking. Methods Mol. Biol. 2012, 819, 221–232. [Google Scholar] [PubMed]
- Torchala, M.; Moal, I.H.; Chaleil, R.A.; Fernandez-Recio, J.; Bates, P.A. Swarmdock: A server for flexible protein-protein docking. Bioinformatics 2013, 29, 807–809. [Google Scholar] [CrossRef] [PubMed]
- Mitra, P.; Pal, D. Prune and probe—Two modular web services for protein-protein docking. Nucleic Acids Res. 2011, 39, W229–W234. [Google Scholar] [CrossRef] [PubMed]
- Lesk, V.I.; Sternberg, M.J. 3D-garden: A system for modelling protein-protein complexes based on conformational refinement of ensembles generated with the marching cubes algorithm. Bioinformatics 2008, 24, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, V.; Moallem, T.M.; Wade, R.C.; Vriend, G.; Edelman, M. CASP2 molecular docking predictions with the LIGIN software. Proteins 1997, 1, 210–214. [Google Scholar] [CrossRef]
- Camacho, C.J.; Gatchell, D.W. Sucessful discrimination of protein interactions. Proteins 2003, 52, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Roberts, V.A.; Thompson, E.E.; Pique, M.E.; Perez, M.S.; Ten Eyck, L.F. Dot2: Macromolecular docking with improved biophysical models. J. Comput. Chem. 2013, 34, 1743–1758. [Google Scholar] [CrossRef] [PubMed]
- Lyskov, S.; Gray, J.J. The rosettadock server for local protein-protein docking. Nucleic Acids Res. 2008, 36, W233–W238. [Google Scholar] [CrossRef] [PubMed]
- Kowalsman, N.; Eisenstein, M. Inherent limitations in protein-protein docking procedures. Bioinformatics 2007, 23, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, D.W.; Venkatraman, V. Ultra-fast fft protein docking on graphics processors. Bioinformatics 2010, 26, 2398–2405. [Google Scholar] [CrossRef] [PubMed]
- Tovchigrechko, A.; Vakser, I.A. Development and testing of an automated approach to protein docking. Proteins 2005, 60, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins 2013, 81, 2159–2166. [Google Scholar] [CrossRef] [PubMed]
- Comeau, S.R.; Gatchell, D.W.; Vajda, S.; Camacho, C.J. Cluspro: An automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 2004, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Kastritis, P.L.; Bonvin, A.M. Are scoring functions in protein-protein docking ready to predict interactomes? Clues from a novel binding affinity benchmark. J. Proteome Res. 2010, 9, 2216–2225. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.; Sacquin-Mora, S.; Dimitrova, V.; Laine, E.; Ponty, Y.; Carbone, A. Protein-protein interactions in a crowded environment: An analysis via cross-docking simulations and evolutionary information. PLoS Comput. Biol. 2013, 9, e1003369. [Google Scholar] [CrossRef] [PubMed]
- Levieux, G.; Tiger, G.; Mader, S.; Zagury, J.F.; Natkin, S.; Montes, M. Udock, the interactive docking entertainment system. Faraday Discuss. 2014, 169, 425–441. [Google Scholar] [CrossRef] [PubMed]
- Andrusier, N.; Nussinov, R.; Wolfson, H.J. Firedock: Fast interaction refinement in molecular docking. Proteins 2007, 69, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Mashiach, E.; Schneidman-Duhovny, D.; Andrusier, N.; Nussinov, R.; Wolfson, H.J. Firedock: A web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008, 36, W229–W232. [Google Scholar] [CrossRef] [PubMed]
- Gabb, H.A.; Jackson, R.M.; Sternberg, M.J. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol. 1997, 272, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Emekli, U.; Schneidman-Duhovny, D.; Wolfson, H.J.; Nussinov, R.; Haliloglu, T. Hingeprot: Automated prediction of hinges in protein structures. Proteins 2008, 70, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Neves, M.A.; Totrov, M.; Abagyan, R. Docking and scoring with icm: The benchmarking results and strategies for improvement. J. Comput. Aided Mol. Des. 2012, 26, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. Patchdock and symmdock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- London, N.; Schueler-Furman, O. Funhunt: Model selection based on energy landscape characteristics. Biochem. Soc. Trans. 2008, 36, 1418–1421. [Google Scholar] [CrossRef] [PubMed]
- Mashiach, E.; Nussinov, R.; Wolfson, H.J. Fiberdock: Flexible induced-fit backbone refinement in molecular docking. Proteins 2010, 78, 1503–1519. [Google Scholar] [PubMed]
- Mashiach, E.; Nussinov, R.; Wolfson, H.J. Fiberdock: A web server for flexible induced-fit backbone refinement in molecular docking. Nucleic Acids Res. 2010, 38, W457–W461. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Zhang, Y. Protein-protein complex structure predictions by multimeric threading and template recombination. Structure 2011, 19, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Guo, L.; Hu, L.; Wang, J. Specificity and affinity quantification of protein-protein interactions. Bioinformatics 2013, 29, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.J.; Zhang, C. Fastcontact: Rapid estimate of contact and binding free energies. Bioinformatics 2005, 21, 2534–2536. [Google Scholar] [CrossRef] [PubMed]
- Chermak, E.; Petta, A.; Serra, L.; Vangone, A.; Scarano, V.; Cavallo, L.; Oliva, R. Consrank: A server for the analysis, comparison and ranking of docking models based on inter-residue contacts. Bioinformatics 2015, 31, 1481–1483. [Google Scholar] [CrossRef] [PubMed]
- Gajda, M.J.; Tuszynska, I.; Kaczor, M.; Bakulina, A.Y.; Bujnicki, J.M. Filtrest3d: Discrimination of structural models using restraints from experimental data. Bioinformatics 2010, 26, 2986–2987. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Li, L.; Weng, Z. Zdock: An initial-stage protein-docking algorithm. Proteins 2003, 52, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Mintseris, J.; Pierce, B.; Wiehe, K.; Anderson, R.; Chen, R.; Weng, Z. Integrating statistical pair potentials into protein complex prediction. Proteins 2007, 69, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Mosca, R.; Pons, C.; Fernandez-Recio, J.; Aloy, P. Pushing structural information into the yeast interactome by high-throughput protein docking experiments. PLoS Comput. Biol. 2009, 5, e1000490. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.; Weng, Z. Zrank: Reranking protein docking predictions with an optimized energy function. Proteins 2007, 67, 1078–1086. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A.M. Haddock: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.J.; van Dijk, A.D.; Krzeminski, M.; van Dijk, M.; Thureau, A.; Hsu, V.; Wassenaar, T.; Bonvin, A.M. Haddock versus haddock: New features and performance of haddock2.0 on the capri targets. Proteins 2007, 69, 726–733. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.J.; Zacharias, M. Attract-em: A new method for the computational assembly of large molecular machines using cryo-em maps. PLoS ONE 2012, 7, e49733. [Google Scholar] [CrossRef] [PubMed]
- May, A.; Zacharias, M. Accounting for global protein deformability during protein-protein and protein-ligand docking. Biochim. Biophys. Acta 2005, 1754, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, M. Protein-protein docking with a reduced protein model accounting for side-chain flexibility. Protein Sci. 2003, 12, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Janin, J.; Wodak, S. The Third Capri Assessment Meeting Toronto, Canada, 20–21 April 2007. Structure 2007, 15, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Anishchenko, I.; Kundrotas, P.J.; Tuzikov, A.V.; Vakser, I.A. Protein models docking benchmark 2. Proteins 2015, 83, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Esmaielbeiki, R.; Nebel, J.C. Scoring docking conformations using predicted protein interfaces. BMC Bioinform. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Fleishman, S.J.; Whitehead, T.A.; Strauch, E.M.; Corn, J.E.; Qin, S.; Zhou, H.X.; Mitchell, J.C.; Demerdash, O.N.; Takeda-Shitaka, M.; Terashi, G.; et al. Community-wide assessment of protein-interface modeling suggests improvements to design methodology. J. Mol. Biol. 2011, 414, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Vreven, T.; Hwang, H.; Weng, Z. Integrating atom-based and residue-based scoring functions for protein-protein docking. Protein Sci. 2011, 20, 1576–1586. [Google Scholar] [CrossRef] [PubMed]
- Brenke, R.; Kozakov, D.; Chuang, G.Y.; Beglov, D.; Hall, D.; Landon, M.R.; Mattos, C.; Vajda, S. Fragment-based identification of druggable “hot spots” of proteins using fourier domain correlation techniques. Bioinformatics 2009, 25, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Hajduk, P.J.; Huth, J.R.; Fesik, S.W. Druggability indices for protein targets derived from nmr-based screening data. J. Med. Chem. 2005, 48, 2518–2525. [Google Scholar] [CrossRef] [PubMed]
- Morrow, J.K.; Zhang, S. Computational prediction of protein hot spot residues. Curr. Pharm. Des. 2012, 18, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Dennis, S.; Kortvelyesi, T.; Vajda, S. Computational mapping identifies the binding sites of organic solvents on proteins. Proc. Natl. Acad. Sci. USA 2002, 99, 4290–4295. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Zhang, Q.C.; Chen, Z.; Meng, Y.; Guan, J.; Zhou, S. PredHS: A web server for predicting protein–protein interaction hot spots by using structural neighborhood properties. Nucleic Acids Res. 2015, 42, W290–W295. [Google Scholar] [CrossRef] [PubMed]
- Lise, S.; Buchan, D.; Pontil, M.; Jones, D.T. Predictions of hot spot residues at protein-protein interfaces using support vector machines. PLoS ONE 2011, 6, e16774. [Google Scholar] [CrossRef] [PubMed]
- Geppert, T.; Hoy, B.; Wessler, S.; Schneider, G. Context-based identification of proteinprotein interfaces and “hot-spot” residues. Chem. Biol. 2011, 18, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Tuncbag, N.; Keksin, O.; Gursoy, A. Hotpoint: Hot spot prediction server for protein interfaces. Nucleic Acids Res. 2010, 38, W402–W406. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Mitchell, J.C. KFC2: A knowledge-based hot spot prediction method based on interface solvation, atomic density and plasticity features. Proteins 2011, 79, 2671–2683. [Google Scholar] [CrossRef] [PubMed]
- Mora, J.S.; Assi, S.A. Presaging Critical Residues in Protein interfaces-Web Server (PCRPi-W): A Web Server to Chart Hot Spots in Protein Interfaces. PLoS ONE 2010, 5, e12352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cesareni, G.; Panni, S.; Nardelli, G.; Castagnoli, L. Can we infer peptide recognition specificity mediated by sh3 domains? FEBS Lett. 2002, 513, 38–44. [Google Scholar] [CrossRef]
- London, N.; Raveh, B.; Cohen, E.; Fathi, G.; Schueler-Furman, O. Rosetta flexpepdock web server—High resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011, 39, W249–W253. [Google Scholar] [CrossRef] [PubMed]
- Madden, D.R.; Gorga, J.C.; Strominger, J.L.; Wiley, D.C. The three-dimensional structure of HLA-b27 at 2.1 Å resolution suggests a general mechanism for tight peptide binding to mhc. Cell 1992, 70, 1035–1048. [Google Scholar] [CrossRef]
- Songyang, Z.; Fanning, A.S.; Fu, C.; Xu, J.; Marfatia, S.M.; Chishti, A.H.; Crompton, A.; Chan, A.C.; Anderson, J.M.; Cantley, L.C. Recognition of unique carboxyl-terminal motifs by distinct pdz domains. Science 1997, 275, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Sudol, M. Structure and function of the ww domain. Prog. Biophys. Mol. Biol. 1996, 65, 113–132. [Google Scholar] [CrossRef]
- Wilson, W.D. Tech.Sight. Analyzing biomolecular interactions. Science 2002, 295, 2103–2105. [Google Scholar] [CrossRef] [PubMed]
- Shuker, S.B.; Hajduk, P.J.; Meadows, R.P.; Fesik, S.W. Discovering high-affinity ligands for proteins: Sar by nmr. Science 1996, 274, 1531–1534. [Google Scholar] [CrossRef] [PubMed]
- Erlanson, D.A. Fragment-based lead discovery: A chemical update. Curr. Opin. Biotechnol. 2006, 17, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, S.; Gullberg, M.; Jarvius, J.; Olsson, C.; Pietras, K.; Gustafsdottir, S.M.; Ostman, A.; Landegren, U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002, 20, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Chu, A.; Li, W.; Wang, B.; Shelton, F.; Otero, F.; Nguyen, D.G.; Caldwell, J.S.; Chen, Y.A. Lipid g protein-coupled receptor ligand identification using β-arrestin pathhunter assay. J. Biol. Chem. 2009, 284, 12328–12338. [Google Scholar] [CrossRef] [PubMed]
- Karpova, T.; McNally, J.G. Detecting protein-protein interactions with cfp-yfp fret by acceptor photobleaching. Curr. Protoc. Cytom. 2006. [Google Scholar] [CrossRef]
- Galdeano, C.; Gadd, M.S.; Soares, P.; Scaffidi, S.; Van Molle, I.; Birced, I.; Hewitt, S.; Dias, D.M.; Ciulli, A. Structure-guided design and optimization of small molecules targeting the protein-protein interaction between the von hippel-lindau (VHL) e3 ubiquitin ligase and the hypoxia inducible factor (HIF) α subunit with in vitro nanomolar affinities. J. Med. Chem. 2014, 57, 8657–8663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mani, T.; Wang, F.; Knabe, W.E.; Sinn, A.L.; Khanna, M.; Jo, I.; Sandusky, G.E.; Sledge, G.W., Jr.; Jones, D.R.; Khanna, R.; et al. Small-molecule inhibition of the uPAR∙uPA interaction: Synthesis, biochemical, cellular, in vivo pharmacokinetics and efficacy studies in breast cancer metastasis. Bioorg. Med. Chem. 2013, 21, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Hain, A.U.; Bartee, D.; Sanders, N.G.; Miller, A.S.; Sullivan, D.J.; Levitskaya, J.; Meyers, C.F.; Bosch, J. Identification of an ATG8-ATG3 protein-protein interaction inhibitor from the medicines for malaria venture malaria box active in blood and liver stage plasmodium falciparum parasites. J. Med. Chem. 2014, 57, 4521–4531. [Google Scholar] [CrossRef] [PubMed]
- Hain, A.U.; Weltzer, R.R.; Hammond, H.; Jayabalasingham, B.; Dinglasan, R.R.; Graham, D.R.; Colquhoun, D.R.; Coppens, I.; Bosch, J. Structural characterization and inhibition of the plasmodium ATG8-ATG3 interaction. J. Struct. Biol. 2012, 180, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Duong-Thi, M.D.; Bergstrom, G.; Mandenius, C.F.; Bergstrom, M.; Fex, T.; Ohlson, S. Comparison of weak affinity chromatography and surface plasmon resonance in determining affinity of small molecules. Anal. Biochem. 2014, 461, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and autodocktools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Ewing, T.J.; Makino, S.; Skillman, A.G.; Kuntz, I.D. Dock 4.0: Search strategies for automated molecular docking of flexible molecule databases. J. Comput. Aided Mol. Des. 2001, 15, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Nussinov, R.; Wolfson, H.J. Automatic prediction of protein interactions with large scale motion. Proteins 2007, 69, 764–773. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Cross, J.B.; Thompson, D.C.; Rai, B.K.; Baber, J.C.; Fan, K.Y.; Hu, Y.; Humblet, C. Comparison of several molecular docking programs: Pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 2009, 49, 1455–1474. [Google Scholar] [CrossRef] [PubMed]
- Saraogi, I.; Hamilton, A.D. α-Helix mimetics as inhibitors of protein-protein interactions. Biochem. Soc. Trans. 2008, 36, 1414–1417. [Google Scholar] [CrossRef] [PubMed]
- Orner, B.P.; Ernst, J.T.; Hamilton, A.D. Toward proteomimetics: Terphenyl derivatives as structural and functional mimics of extended regions of an α-helix. J. Am. Chem. Soc. 2001, 123, 5382–5383. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.M.; Nevola, L.; Ross, N.T.; Lee, G.I.; Hamilton, A.D. Synthetic inhibitors of extended helix-protein interactions based on a biphenyl 4,4′-dicarboxamide scaffold. Chembiochem 2009, 10, 829–833. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.H.; Waldman, A.S.; Nowick, J.S. Two new β-strand mimics. Bioorg. Med. Chem. 1999, 7, 29–38. [Google Scholar] [CrossRef]
- Fasan, R.; Dias, R.L.; Moehle, K.; Zerbe, O.; Vrijbloed, J.W.; Obrecht, D.; Robinson, J.A. Using a β-hairpin to mimic an α-helix: Cyclic peptidomimetic inhibitors of the p53-hdm2 protein-protein interaction. Angew. Chem. 2004, 43, 2109–2112. [Google Scholar] [CrossRef] [PubMed]
- Hirschmann, R.F.; Nicolaou, K.C.; Angeles, A.R.; Chen, J.S.; Smith, A.B., 3rd. The β-d-glucose scaffold as a β-turn mimetic. Acc. Chem. Res. 2009, 42, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Ball, L.J.; Kuhne, R.; Schneider-Mergener, J.; Oschkinat, H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angew. Chem. 2005, 44, 2852–2869. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Khoury, K.; Chanas, T.; Dömling, A. Multicomponent Synthesis of Diverse 1,4-Benzodiazepine Scaffolds. Org. Lett. 2012, 14, 5916–5919. [Google Scholar] [CrossRef] [PubMed]
- Ko, E.; Liu, J.; Burgess, K. Minimalist and universal peptidomimetics. Chem. Soc. Rev. 2011, 40, 4411–4421. [Google Scholar] [CrossRef] [PubMed]
- Jubb, H.; Higueruelo, A.P.; Winter, A.; Blundell, T.L. Structural biology and drug discovery for protein-protein interactions. Trends Pharmacol. Sci. 2012, 33, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Oltersdorf, T.; Elmore, S.W.; Shoemaker, A.R.; Armstrong, R.C.; Augeri, D.J.; Belli, B.A.; Bruncko, M.; Deckwerth, T.L.; Dinges, J.; Hajduk, P.J.; et al. An inhibitor of bcl-2 family proteins induces regression of solid tumours. Nature 2005, 435, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Hiroaki, H. Recent applications of isotopic labeling for protein nmr in drug discovery. Expert Opin. Drug Discov. 2013, 8, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.K.; Gellman, S.H. Targeting protein-protein interactions: Lessons from P53/MDM2. Biopolymers 2007, 88, 657–686. [Google Scholar] [CrossRef] [PubMed]
- Rechfeld, F.; Gruber, P.; Hofmann, J.; Kirchmair, J. Modulators of protein-protein interactions: Novel approaches in targeting protein kinases and other pharmaceutically relevant biomolecules. Curr. Top. Med. Chem. 2011, 11, 1305–1319. [Google Scholar] [PubMed]
- Fedosyuk, S.; Grishkovskaya, I.; de Almeida Ribeiro, E., Jr.; Skern, T. Characterization and structure of the vaccinia virus nf-kappab antagonist a46. J. Biol. Chem. 2014, 289, 3749–3762. [Google Scholar] [CrossRef] [PubMed]
- Dhruv, H.; Loftus, J.C.; Narang, P.; Petit, J.L.; Fameree, M.; Burton, J.; Tchegho, G.; Chow, D.; Yin, H.; Al-Abed, Y.; et al. Structural basis and targeting of the interaction between fibroblast growth factor-inducible 14 and tumor necrosis factor-like weak inducer of apoptosis. J. Biol. Chem. 2013, 288, 32261–32276. [Google Scholar] [CrossRef] [PubMed]
- Svajger, U.; Brus, B.; Turk, S.; Sova, M.; Hodnik, V.; Anderluh, G.; Gobec, S. Novel toll-like receptor 4 (TLR4) antagonists identified by structure- and ligand-based virtual screening. Eur. J. Med. Chem. 2013, 70, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Isvoran, A.; Badel, A.; Craescu, C.T.; Miron, S.; Miteva, M.A. Exploring nmr ensembles of calcium binding proteins: Perspectives to design inhibitors of protein-protein interactions. BMC Struct. Biol. 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Pala, D.; Castelli, R.; Incerti, M.; Russo, S.; Tognolini, M.; Giorgio, C.; Hassan-Mohamed, I.; Zanotti, I.; Vacondio, F.; Rivara, S.; et al. Combining ligand- and structure-based approaches for the discovery of new inhibitors of the EPHA2-ephrin-A1 interaction. J. Chem. Inf. Model. 2014, 54, 2621–2626. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Kaneko, O.; Thongkukiatkul, A.; Tachibana, M.; Otsuki, H.; Gao, Q.; Tsuboi, T.; Torii, M. Rhoptry neck protein RON2 forms a complex with microneme protein ama1 in plasmodium falciparum merozoites. Parasitol. Int. 2009, 58, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, P.; Beatty, W.L.; Diouf, A.; Herrera, R.; Ambroggio, X.; Moch, J.K.; Tyler, J.S.; Narum, D.L.; Pierce, S.K.; Boothroyd, J.C.; et al. Binding of plasmodium merozoite proteins ron2 and ama1 triggers commitment to invasion. Proc. Natl. Acad. Sci. USA 2011, 108, 13275–13280. [Google Scholar] [CrossRef] [PubMed]
- Devine, S.M.; May, L.T.; Scammells, P.J. Design, synthesis and evaluation of N6-substituted 2-aminoadenosine-5[prime or minute]-N-methylcarboxamides as A3 adenosine receptor agonists. MedChemComm 2014, 5, 192–196. [Google Scholar] [CrossRef]
- Pihan, E.; Delgadillo, R.F.; Tonkin, M.L.; Pugniere, M.; Lebrun, M.; Boulanger, M.J.; Douguet, D. Computational and biophysical approaches to protein-protein interaction inhibition of plasmodium falciparum ama1/ron2 complex. J. Comput. Aided Mol. Des. 2015, 29, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Korb, O.; Stutzle, T.; Exner, T.E. Empirical scoring functions for advanced protein-ligand docking with plants. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Broos, K.; Trekels, M.; Jose, R.A.; Demeulemeester, J.; Vandenbulcke, A.; Vandeputte, N.; Venken, T.; Egle, B.; De Borggraeve, W.M.; Deckmyn, H.; et al. Identification of a small molecule that modulates platelet glycoprotein ib-von willebrand factor interaction. J. Biol. Chem. 2012, 287, 9461–9472. [Google Scholar] [CrossRef] [PubMed]
- Fontayne, A.; Meiring, M.; Lamprecht, S.; Roodt, J.; Demarsin, E.; Barbeaux, P.; Deckmyn, H. The humanized anti-glycoprotein ib monoclonal antibody h6B4-Fab is a potent and safe antithrombotic in a high shear arterial thrombosis model in baboons. Thromb. Haemost. 2008, 100, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Ulrichts, H.; Silence, K.; Schoolmeester, A.; de Jaegere, P.; Rossenu, S.; Roodt, J.; Priem, S.; Lauwereys, M.; Casteels, P.; Van Bockstaele, F.; et al. Antithrombotic drug candidate alx-0081 shows superior preclinical efficacy and safety compared with currently marketed antiplatelet drugs. Blood 2011, 118, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Dumas, J.J.; Kumar, R.; McDonagh, T.; Sullivan, F.; Stahl, M.L.; Somers, W.S.; Mosyak, L. Crystal structure of the wild-type von willebrand factor A1-glycoprotein Ibα complex reveals conformation differences with a complex bearing von willebrand disease mutations. J. Biol. Chem. 2004, 279, 23327–23334. [Google Scholar] [CrossRef] [PubMed]
- Brady, G.P., Jr.; Stouten, P.F. Fast prediction and visualization of protein binding pockets with pass. J. Comput. Aided Mol. Des. 2000, 14, 383–401. [Google Scholar] [CrossRef] [PubMed]
- Pettit, F.K.; Bare, E.; Tsai, A.; Bowie, J.U. Hotpatch: A statistical approach to finding biologically relevant features on protein surfaces. J. Mol. Biol. 2007, 369, 863–879. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Flies, D.B. Molecular mechanisms of t cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 2013, 13, 227–242. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.J.; Ikemizu, S.; Evans, E.J.; Fugger, L.; Bakker, T.R.; van der Merwe, P.A. The nature of molecular recognition by t cells. Nat. Immunol. 2003, 4, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayanajois, S.D. Cell adhesion molecules: Structure, function, drug design, and biomaterials. Curr. Pharm. Des. 2008, 14, 2126–2127. [Google Scholar] [CrossRef] [PubMed]
- Raychaudhuri, S.; Thomson, B.P.; Remmers, E.F.; Eyre, S.; Hinks, A.; Guiducci, C.; Catanese, J.J.; Xie, G.; Stahl, E.A.; Chen, R.; et al. Genetic variants at CD28, prdm1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet. 2009, 41, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
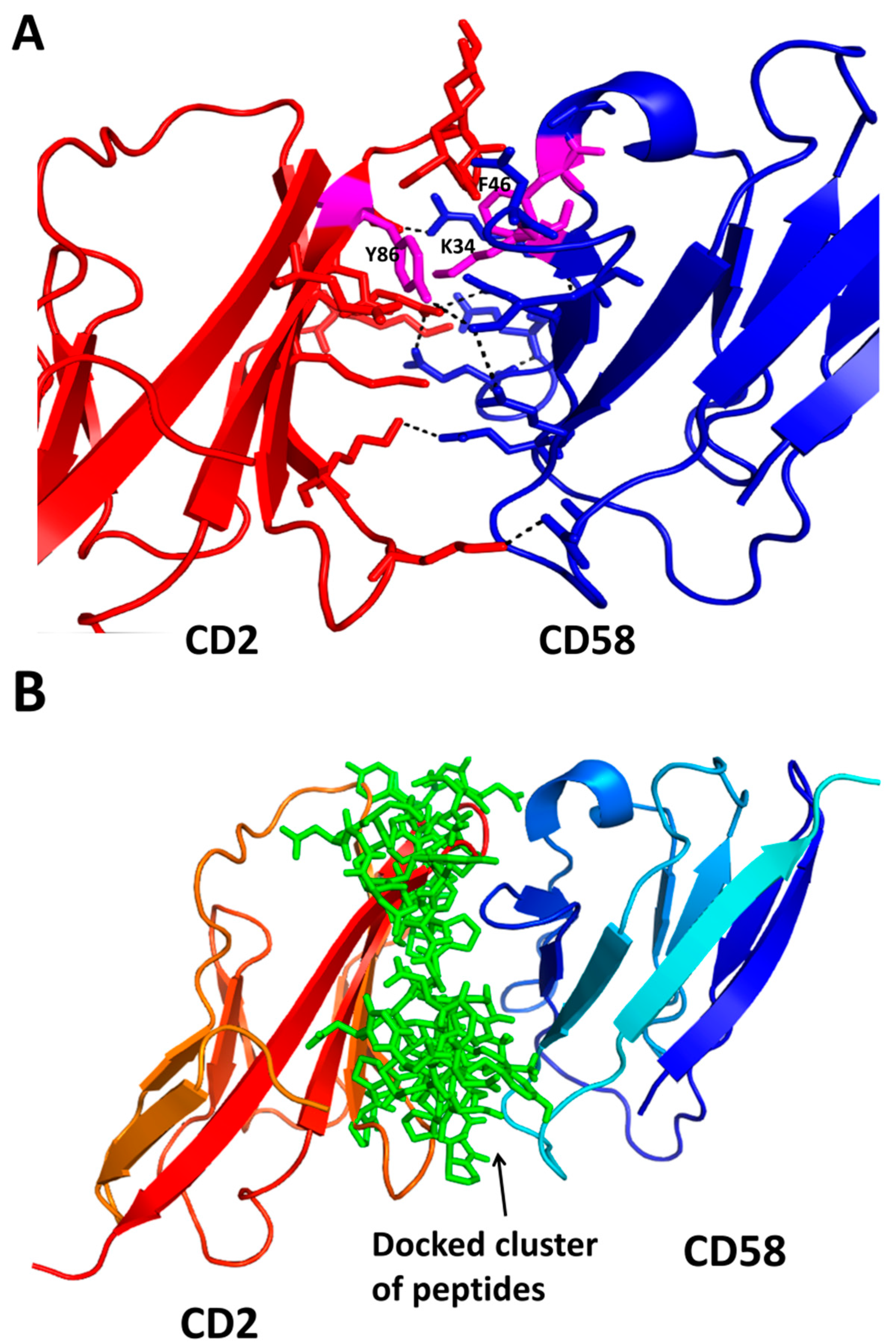
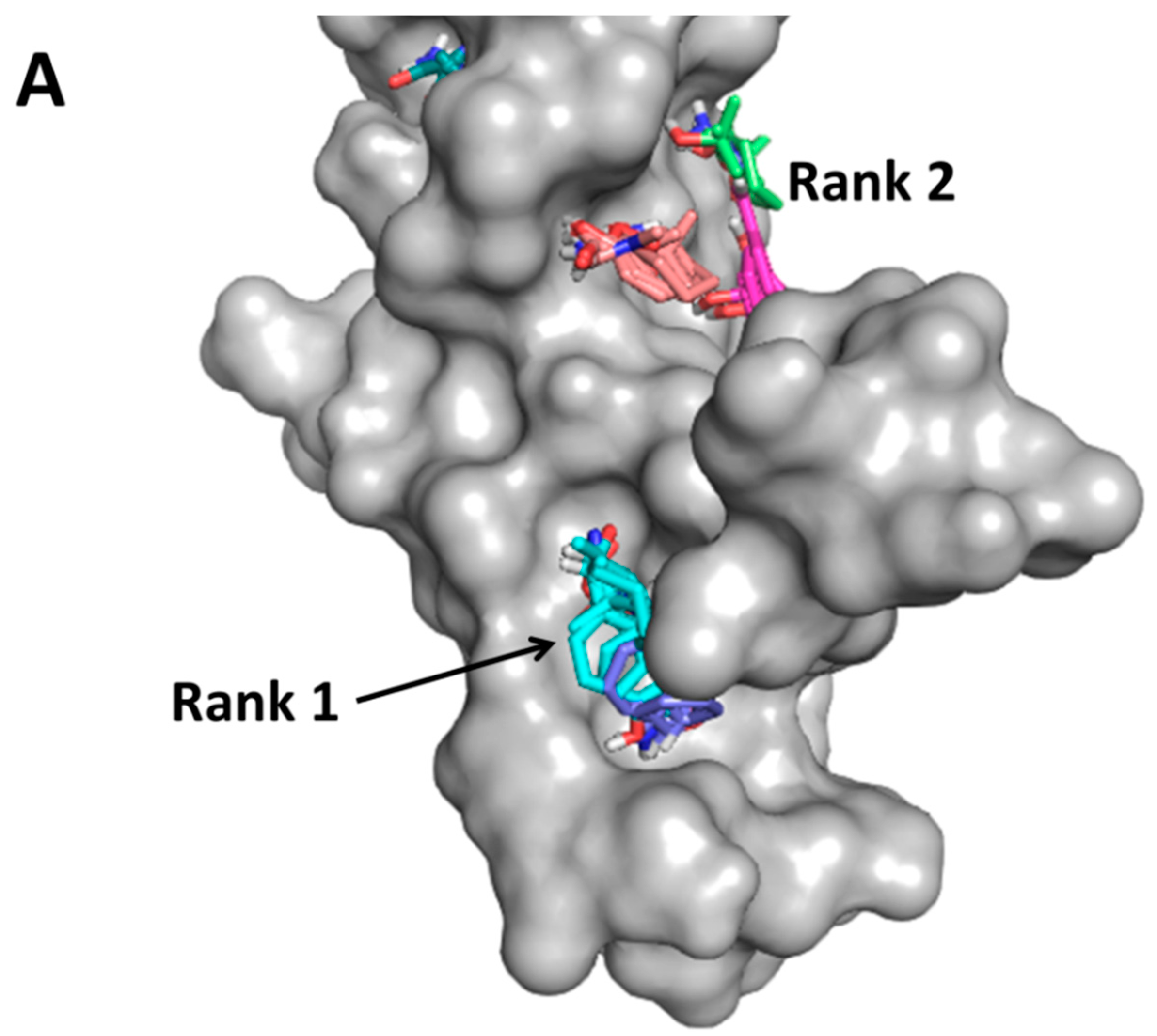
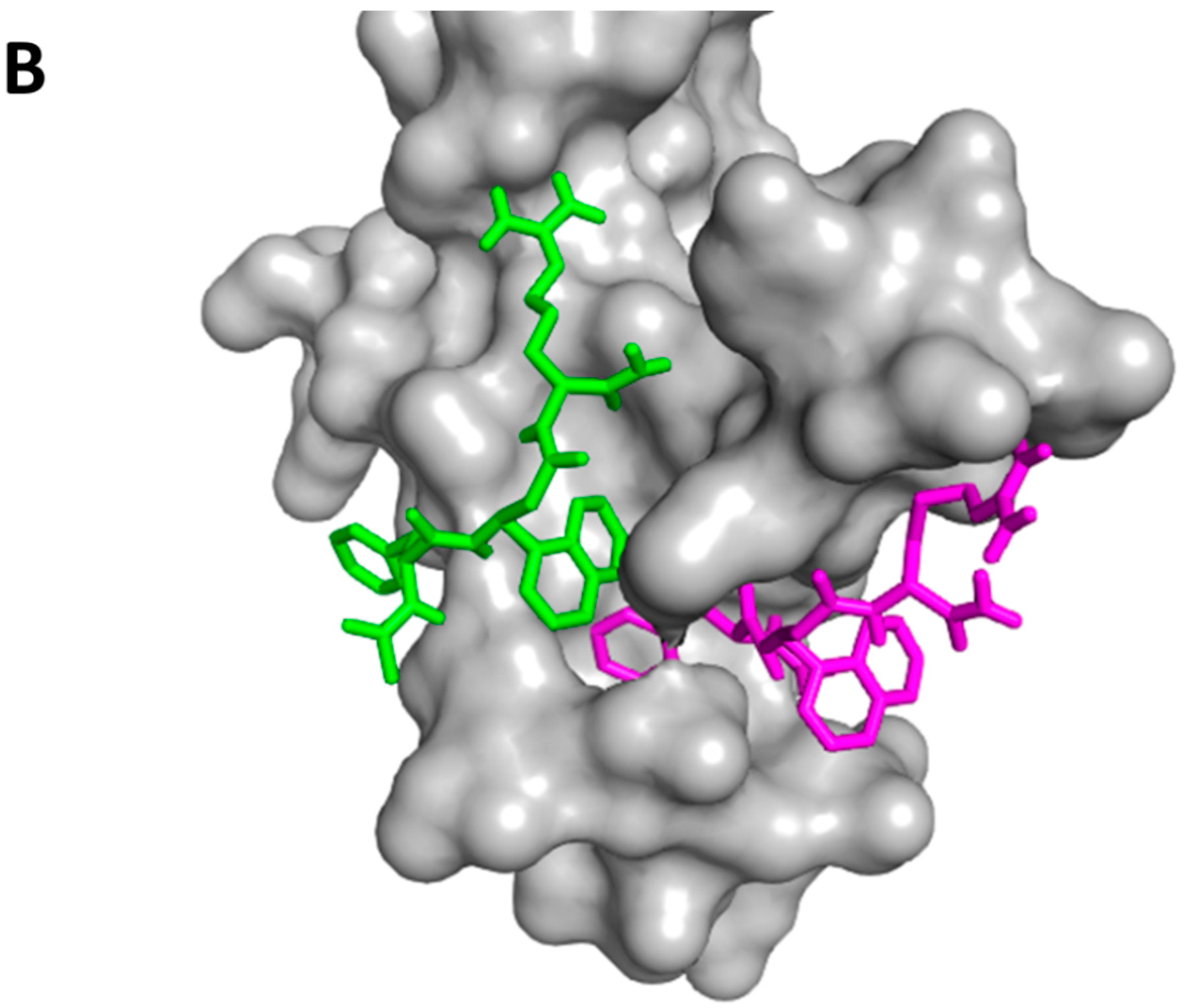
- Kim, M.; Sun, Z.Y.; Byron, O.; Campbell, G.; Wagner, G.; Wang, J.; Reinherz, E.L. Molecular dissection of the CD2-CD58 counter-receptor interface identifies CD2 Tyr86 and CD58 Lys34 residues as the functional “hot spot”. J. Mol. Biol. 2001, 312, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Smolyar, A.; Tan, K.; Liu, J.H.; Kim, M.; Sun, Z.Y.; Wagner, G.; Reinherz, E.L. Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors. Cell 1999, 97, 791–803. [Google Scholar] [CrossRef]
- Giddu, S.; Subramanian, V.; Yoon, H.S.; Satyanarayanajois, S.D. Design of β-hairpin peptides for modulation of cell adhesion by β-turn constraint. J. Med. Chem. 2009, 52, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, A.; Kanthala, S.; Latendresse, J.; Taneja, V.; Satyanarayanajois, S. Immunosuppression by co-stimulatory molecules: Inhibition of CD2-CD48/CD58 interaction by peptides from CD2 to suppress progression of collagen-induced arthritis in mice. Chem. Biol. Drug Des. 2013, 82, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, A.; Weldeghiorghis, T.K.; Taneja, V.; Satyanarayanajois, S.D. Conformationally constrained peptides from CD2 to modulate protein-protein interactions between CD2 and CD58. J. Med. Chem. 2011, 54, 5307–5319. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayanajois, S.D.; Buyuktimkin, B.; Gokhale, A.; Ronald, S.; Siahaan, T.J.; Latendresse, J.R. A peptide from the β-strand region of CD2 protein that inhibits cell adhesion and suppresses arthritis in a mouse model. Chem. Biol. Drug Des. 2010, 76, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, K.M. Structure-based view of epidermal growth factor receptor regulation. Annu. Rev. Biophys. 2008, 37, 353–373. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, R. Her2 therapy. Her2 (ERBB2): Functional diversity from structurally conserved building blocks. Breast Cancer Res. BCR 2007, 9, 202. [Google Scholar] [CrossRef] [PubMed]
- Nahta, R.; Esteva, F.J. Her-2-targeted therapy: Lessons learned and future directions. Clin. Cancer Res. 2003, 9, 5078–5084. [Google Scholar] [PubMed]
- Chang, J.C. Her2 inhibition: From discovery to clinical practice. Clin. Cancer Res. 2007, 13, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Banappagari, S.; Corti, M.; Pincus, S.; Satyanarayanajois, S. Inhibition of protein-protein interaction of HER2-EGFR and HER2-HER3 by a rationally designed peptidomimetic. J. Biomol. Struct. Dyn. 2012, 30, 594–606. [Google Scholar] [CrossRef] [PubMed]
- Banappagari, S.; McCall, A.; Fontenot, K.; Vicente, M.G.; Gujar, A.; Satyanarayanajois, S. Design, synthesis and characterization of peptidomimetic conjugate of bodipy targeting HER2 protein extracellular domain. Eur. J. Med. Chem. 2013, 65, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Banappagari, S.; Ronald, S.; Satyanarayanajois, S.D. A conformationally constrained peptidomimetic binds to the extracellular region of HER2 protein. J. Biomol. Struct. Dyn. 2010, 28, 289–308. [Google Scholar] [CrossRef] [PubMed]
- Banappagari, S.; Ronald, S.; Satyanarayanajois, S.D. Structure-activity relationship of conformationally constrained peptidomimetics for antiproliferative activity in HER2-overexpressing breast cancer cell lines. MedChemComm 2011, 2, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Kanthala, S.; Banappagari, S.; Gokhale, A.; Liu, Y.Y.; Xin, G.; Zhao, Y.; Jois, S. Novel peptidomimetics for inhibition of HER2:HER3 heterodimerization in HER2-positive breast cancer. Chem. Biol. Drug Des. 2014, 85, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Kanthala, S.; Gauthier, T.; Satyanarayanajois, S. Structure-activity relationships of peptidomimetics that inhibit ppi of HER2-HER3. Biopolymers 2014, 101, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayanajois, S.; Villalba, S.; Jianchao, L.; Lin, G.M. Design, synthesis, and docking studies of peptidomimetics based on HER2-herceptin binding site with potential antiproliferative activity against breast cancer cell lines. Chem. Biol. Drug Des. 2009, 74, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.D.; Ultsch, M.; Lingel, A.; Schaefer, G.; Shao, L.; Birtalan, S.; Sidhu, S.S.; Eigenbrot, C. Structure of the complex between HER2 and an antibody paratope formed by side chains from tryptophan and serine. J. Mol. Biol. 2010, 402, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Casey, F.P.; Pihan, E.; Shields, D.C. Discovery of small molecule inhibitors of protein-protein interactions using combined ligand and target score normalization. J. Chem. Inf. Model. 2009, 49, 2708–2717. [Google Scholar] [CrossRef] [PubMed]
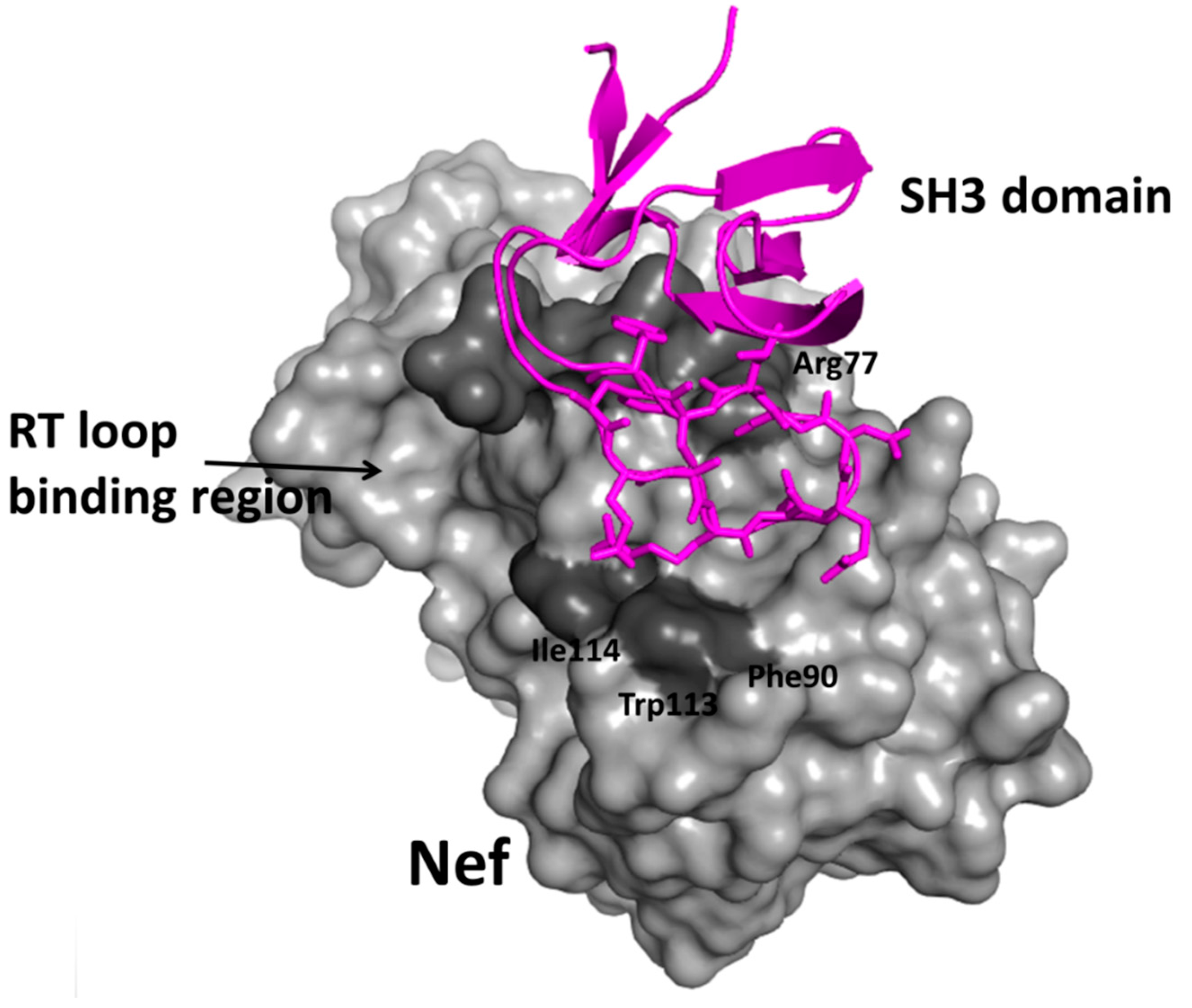
- Betzi, S.; Restouin, A.; Opi, S.; Arold, S.T.; Parrot, I.; Guerlesquin, F.; Morelli, X.; Collette, Y. Protein protein interaction inhibition (2P2I) combining high throughput and virtual screening: Application to the HIV-1 nef protein. Proc. Natl. Acad. Sci. USA 2007, 104, 19256–19261. [Google Scholar] [CrossRef] [PubMed]
- Nomme, J.; Renodon-Corniere, A.; Asanomi, Y.; Sakaguchi, K.; Stasiak, A.Z.; Stasiak, A.; Norden, B.; Tran, V.; Takahashi, M. Design of potent inhibitors of human rad51 recombinase based on brc motifs of brca2 protein: Modeling and experimental validation of a chimera peptide. J. Med. Chem. 2010, 53, 5782–5791. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Hamilton, A.D. Strategies for targeting protein-protein interactions with synthetic agents. Angew. Chem. 2005, 44, 4130–4163. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Bower, K.E.; Beuscher, A.E.; Zhou, B.; Bobkov, A.A.; Olson, A.J.; Vogt, P.K. Stabilizers of the max homodimer identified in virtual ligand screening inhibit myc function. Mol. Pharmacol. 2009, 76, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.K.; Burley, S.K. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 2003, 112, 193–205. [Google Scholar] [CrossRef]
- Orry, A.J.; Abagyan, R.A.; Cavasotto, C.N. Structure-based development of target-specific compound libraries. Drug Discov. Today 2006, 11, 261–266. [Google Scholar] [CrossRef]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sable, R.; Jois, S. Surfing the Protein-Protein Interaction Surface Using Docking Methods: Application to the Design of PPI Inhibitors. Molecules 2015, 20, 11569-11603. https://doi.org/10.3390/molecules200611569
Sable R, Jois S. Surfing the Protein-Protein Interaction Surface Using Docking Methods: Application to the Design of PPI Inhibitors. Molecules. 2015; 20(6):11569-11603. https://doi.org/10.3390/molecules200611569
Chicago/Turabian StyleSable, Rushikesh, and Seetharama Jois. 2015. "Surfing the Protein-Protein Interaction Surface Using Docking Methods: Application to the Design of PPI Inhibitors" Molecules 20, no. 6: 11569-11603. https://doi.org/10.3390/molecules200611569