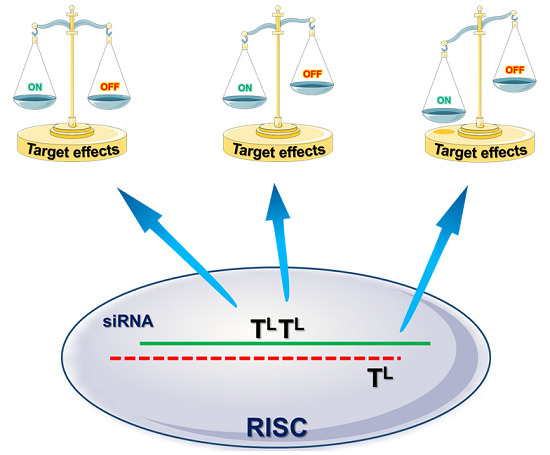
Modulation of the RNA Interference Activity Using Central Mismatched siRNAs and Acyclic Threoninol Nucleic Acids (aTNA) Units
Abstract
:1. Introduction
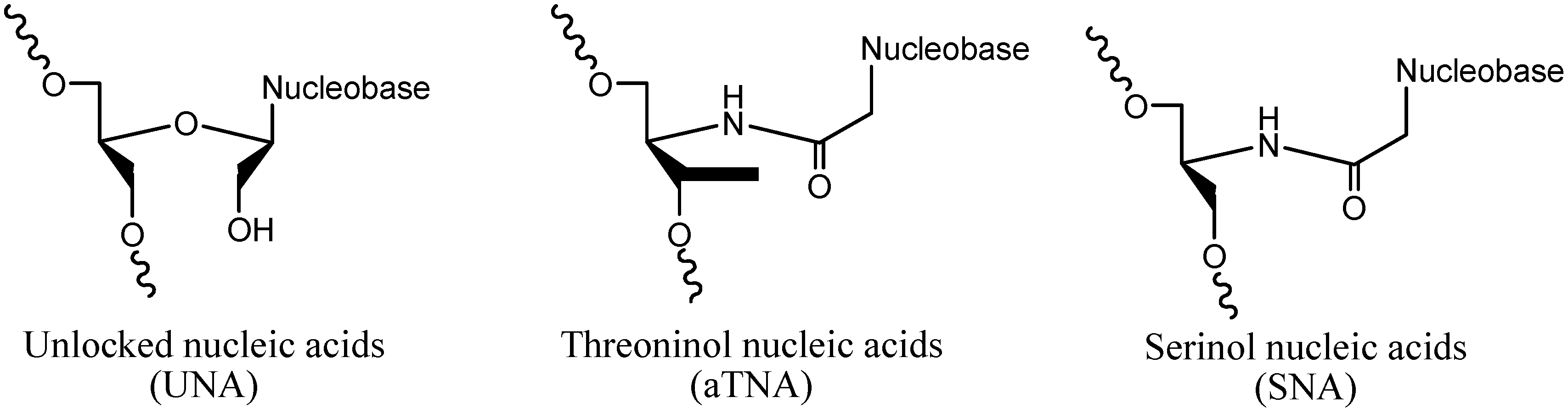
2. Results and Discussion
2.1. Design and Thermodynamic Properties of siRNA Carrying L-Threoninol Monomers
| Antisense ..ZW.. | Sense ..XY.. | IC50 (pM) ± SD | Tm (°C) ± SD | ΔTm (wt) | ΔTm (Parent) | |
|---|---|---|---|---|---|---|
| wt | ..U.. | ..A.. | 9.6 ± 0.5 | 67.8 ± 0.3 | -- | -- |
| wtU9 | ..U.. | ..A.. | 15.4 ± 0.7 | 60.4 ± 0.2 | 7.6 | 7.6 |
| wtC9 | ..U.. | ..A.. | 30.7 ± 0.6 | 59.5 ± 0.4 | 8.5 | 8.5 |
| wtG9 | ..U.. | ..A.. | 15.8 ± 0.2 | 65.9 ± 0.2 | 2.1 | 2.1 |
| wtU10 | ..U.. | ..A.. | No active | 58.2 ± 0.4 | 9.6 | 9.6 |
| wtC10 | ..U.. | ..A.. | No active | 60.0 ± 0.1 | 7.8 | 7.8 |
| wtG10 | ..U.. | ..A.. | 101 ± 0.7 | 64.5 ± 0.6 | 3.5 | 3.5 |
| T10A10 | ..U.. | ..A.. | 111 ± 0.8 | 58.3 ± 0.3 | 9.5 | -- |
| T11A9 | ..U.. | ..A.. | 20.2 ± 0.6 | 59.1 ± 0.3 | 8.7 | -- |
| T10U10 | ..U.. | ..A.. | 216 ± 0.6 | 55.2 ± 0.1 | 12.6 | 3.1 |
| T10C10 | ..U.. | ..A.. | 277 ± 0.9 | 54.1 ± 0.4 | 13.7 | 4.2 |
| T10G10 | ..U.. | ..A.. | 110 ± 0.9 | 55.7 ± 0.3 | 10.8 | 1.3 |
| T11U9 | ..U.. | ..A.. | 35.6 ± 0.4 | 56.4 ± 0.2 | 11.4 | 2.7 |
| T11C9 | ..U.. | ..A.. | 57.3 ± 0.5 | 55.6 ± 0.2 | 12.2 | 3.5 |
| T11G9 | ..U.. | ..A.. | 26.5 ± 0.8 | 58.4 ± 0.5 | 9.4 | 0.7 |

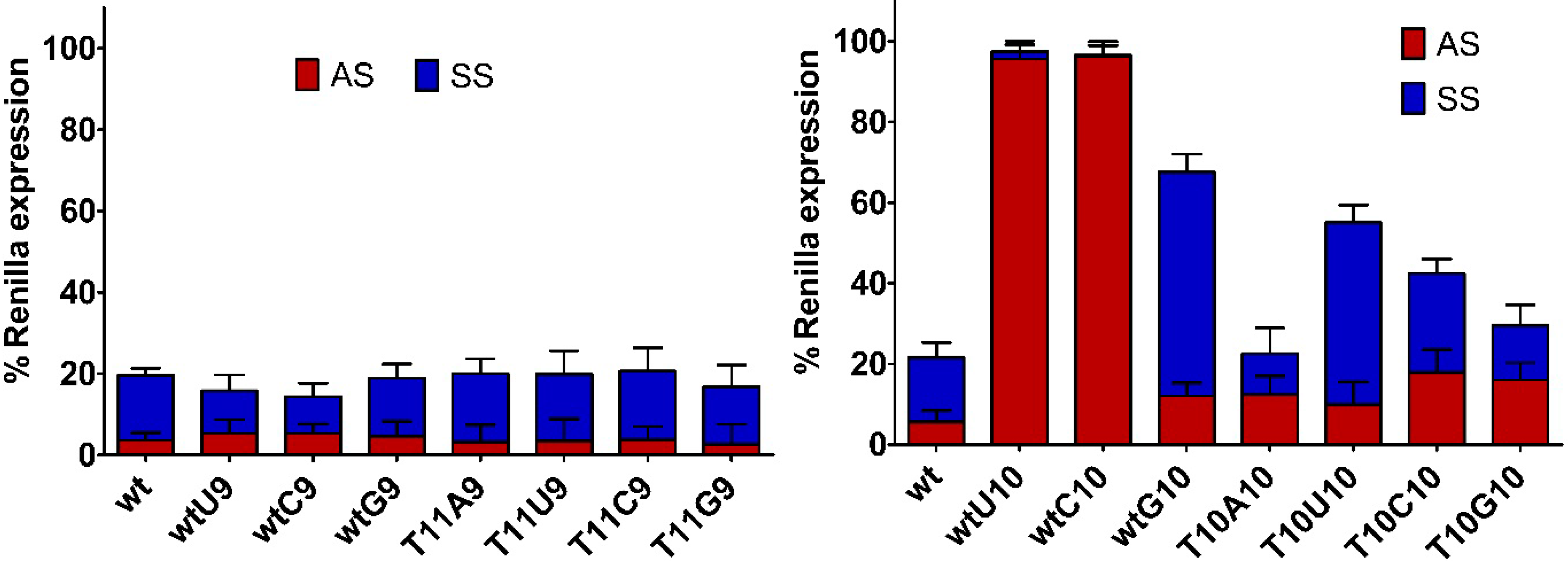
2.2. Impact of Mismatches and/or L-Threoninol Modifications on the Silencing Activity of siRNAs
2.3. Central Mismatched siRNA: Trying to Bias the Silence
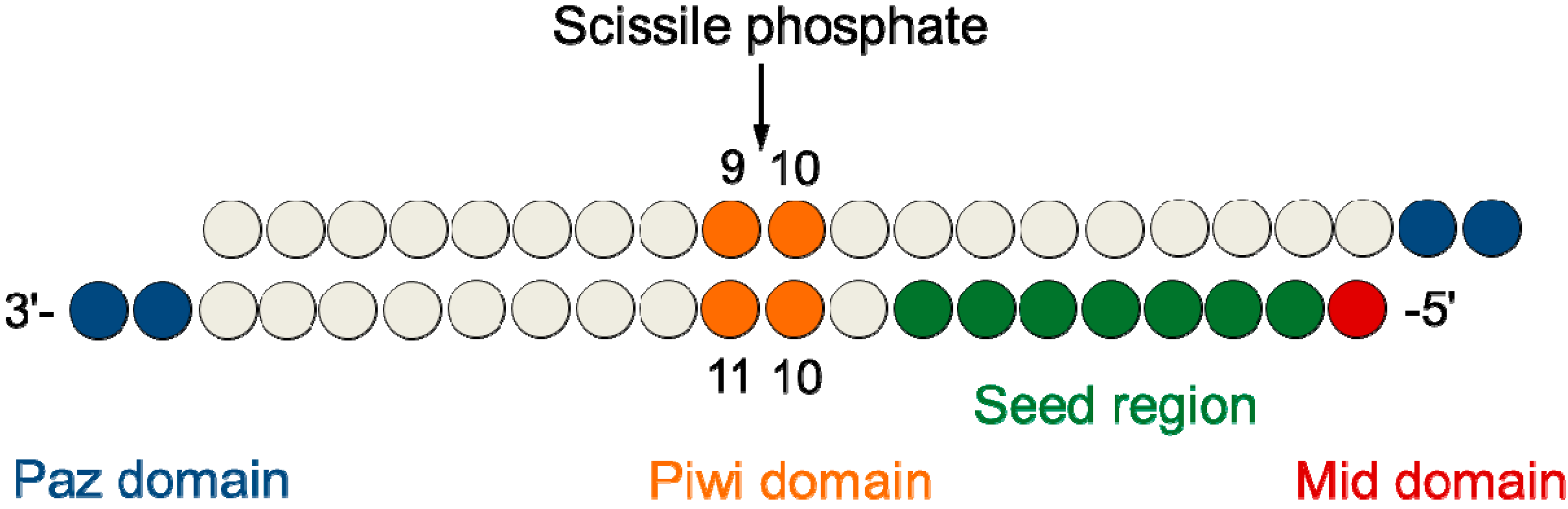
2.4. Central L-Threoninol Modified siRNAs act through an Ago2-Mediated Mechanism
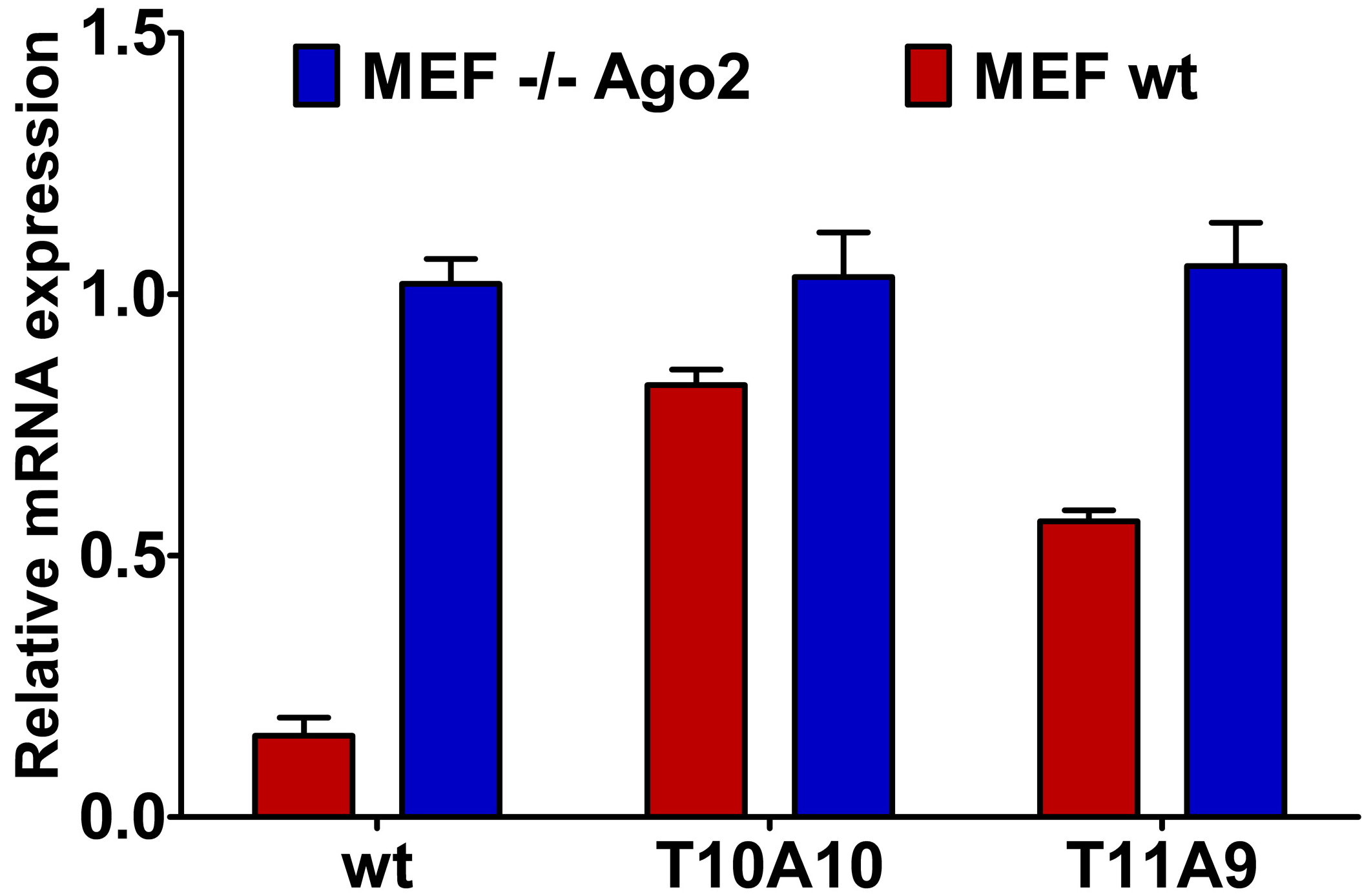
2.5. Single-Stranded siRNAs Experiments
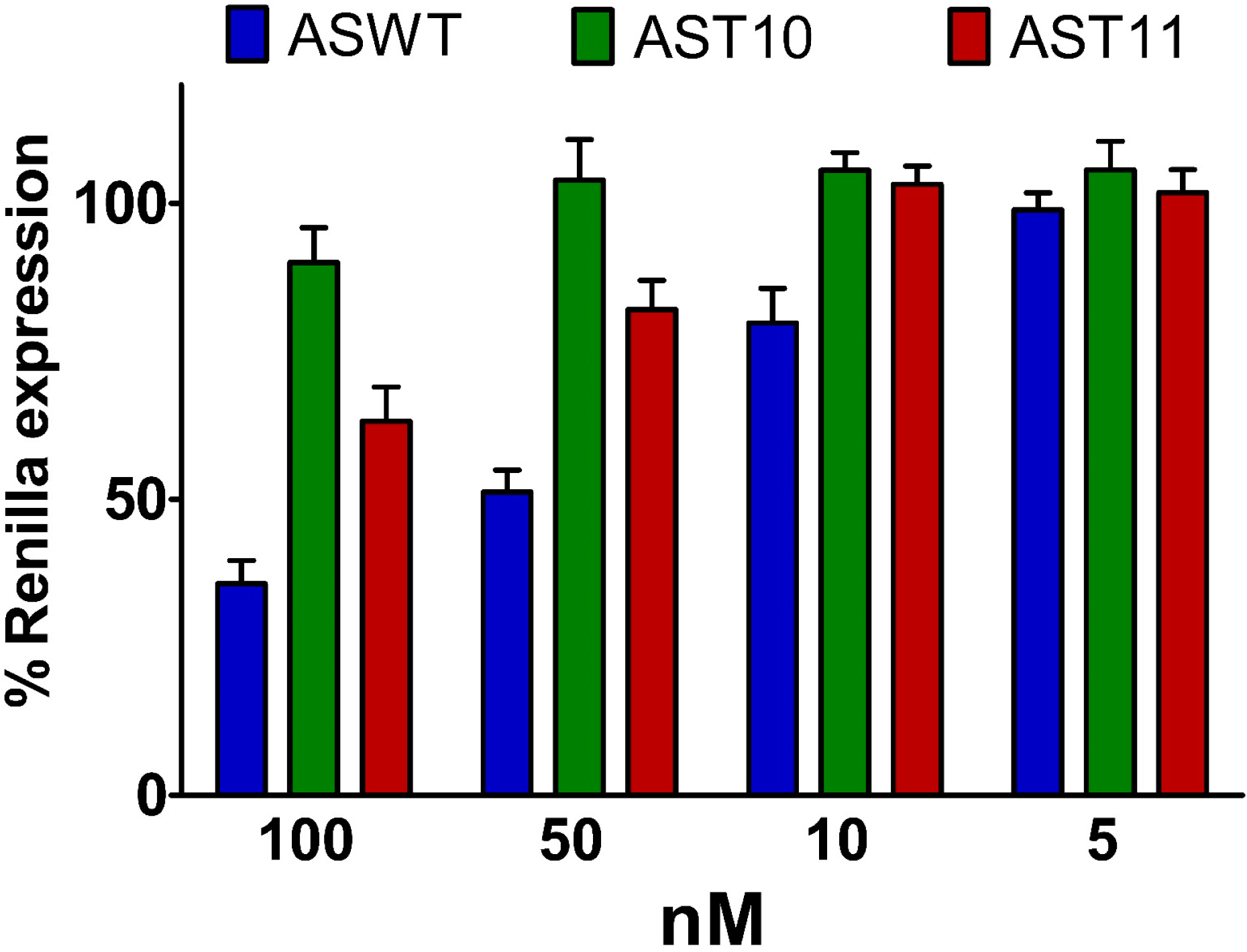
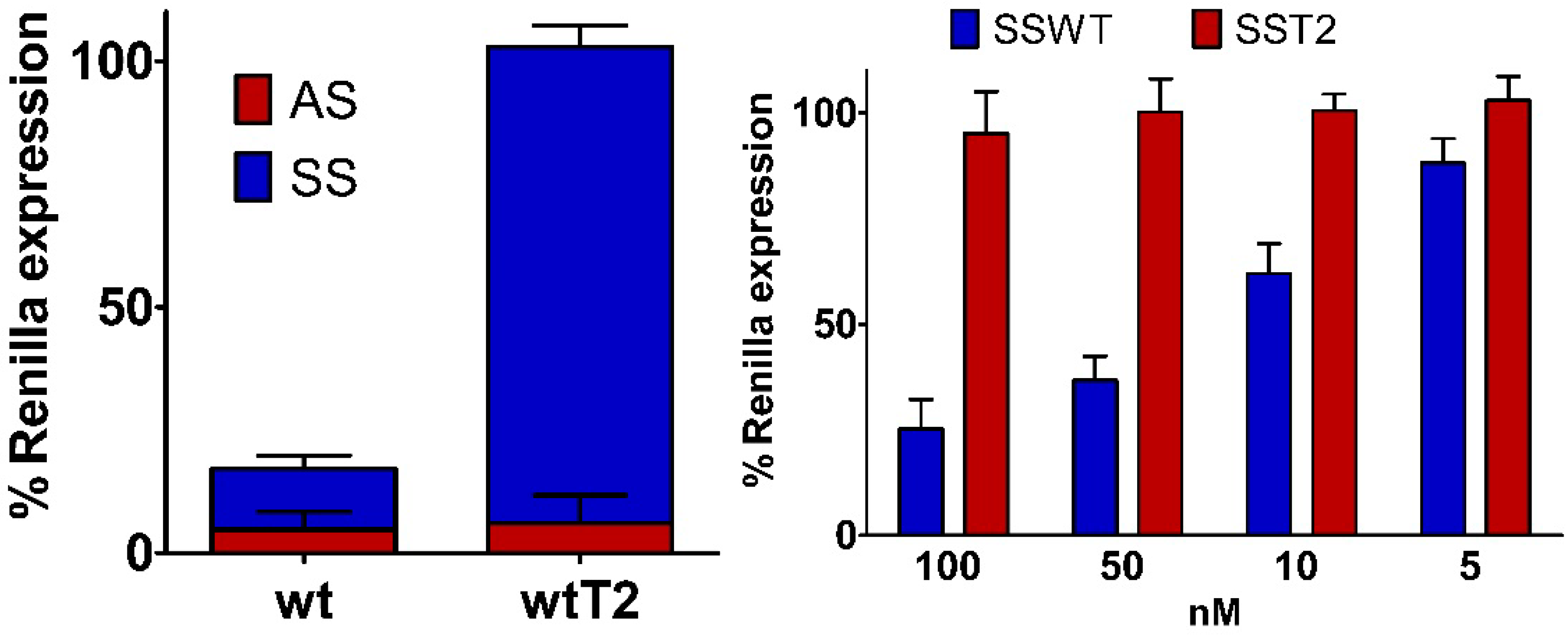
2.6. Silencing Asymmetry and “Strand-Blocking” Effect of TL Modification

| Antisense ..A.. | Sense ..K.. | IC50 (pM) ± SD | Tm (°C) ± SD | Δ Tm (wt) | |
|---|---|---|---|---|---|
| wt | ..A.. | ..U.. | 9.6 ± 0.5 | 67.8 ± 0.3 | -- |
| wtT2 | ..A.. | ..TL.. | 13.4 ± 0.3 | 66.7 ± 0.3 | 1.1 |
3. Experimental Section
3.1. RNA Synthesis
3.2. Deprotection and Purification of Unmodified and Modified RNA Oligonucleotide
3.3. SiRNA Preparation
3.4. Thermal Denaturation Studies
3.5. Cells
3.6. PsiCHECK2 on-/off-Target Reporters
3.7. Transfection and Luciferase Assay
3.8. Ago2-Mediated Silencing Assay
3.9. Single-Stranded siRNA 5'-End Phosphorylation
3.10. Isolation of RNA and RT-qPCR
3.11. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Burnett, J.C.; Rossi, J.J. RNA-based therapeutics: Current progress and future prospects. Chem. Biol. 2012, 19, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Martinez, T.; Wright, N.; Lopez-Fraga, M.; Jimenez, A.I.; Paneda, C. Silencing human genetic diseases with oligonucleotide-based therapies. Hum. Genet. 2013, 132, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Grijalvo, S.; Avino, A.; Eritja, R. Oligonucleotide delivery: A patent review (2010–2013). Expert Opin. Ther. Pat. 2014, 24, 801–819. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, C.C.; Choy, K.W.; Du, Q.; Chen, J.; Wang, Q.; Li, L.; Chung, T.K.; Tang, T. Therapeutic potentials of gene silencing by RNA interference: Principles, challenges, and new strategies. Gene 2014, 538, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Judge, A.D.; Sood, V.; Shaw, J.R.; Fang, D.; McClintock, K.; MacLachlan, I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005, 23, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Deleavey, G.F.; Damha, M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012, 19, 937–954. [Google Scholar] [CrossRef] [PubMed]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [PubMed]
- Shukla, S.; Sumaria, C.S.; Pradeepkumar, P.I. Exploring chemical modifications for siRNA therapeutics: A structural and functional outlook. ChemMedChem 2010, 5, 328–349. [Google Scholar] [CrossRef] [PubMed]
- Murayama, K.; Tanaka, Y.; Toda, T.; Kashida, H.; Asanuma, H. Highly stable duplex formation by artificial nucleic acids acyclic threoninol nucleic acid (aTNA) and serinol nucleic acid (SNA) with acyclic scaffolds. Chemistry 2013, 19, 14151–14158. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Nielsen, C.B.; Nielsen, K.E.; Jensen, G.A.; Bondensgaard, K.; Singh, S.K.; Rajwanshi, V.K.; Koshkin, A.A.; Dahl, B.M.; Wengel, J.; et al. The conformations of locked nucleic acids (LNA). J. Mol. Recognit. 2000, 13, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Elmen, J.; Thonberg, H.; Ljungberg, K.; Frieden, M.; Westergaard, M.; Xu, Y.; Wahren, B.; Liang, Z.; Orum, H.; Koch, T.; et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005, 33, 439–447. [Google Scholar] [CrossRef]
- Terrazas, M.; Ocampo, S.M.; Perales, J.C.; Marquez, V.E.; Eritja, R. Effect of north bicyclo[3.1.0]hexane 2'-deoxy-pseudosugars on RNA interference: A novel class of siRNA modification. ChemBioChem 2011, 12, 1056–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anzahaee, M.Y.; Deleavey, G.F.; Le, P.U.; Fakhoury, J.; Petrecca, K.; Damha, M.J. Arabinonucleic acids: 2'-stereoisomeric modulators of siRNA activity. Nucleic Acid Ther. 2014, 24, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Dowler, T.; Bergeron, D.; Tedeschi, A.L.; Paquet, L.; Ferrari, N.; Damha, M.J. Improvements in siRNA properties mediated by 2'-deoxy-2'-fluoro-beta-d-arabinonucleic acid (FANA). Nucleic Acids Res. 2006, 34, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Laursen, M.B.; Pakula, M.M.; Gao, S.; Fluiter, K.; Mook, O.R.; Baas, F.; Langklaer, N.; Wengel, S.L.; Wengel, J.; Kjems, J.; et al. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol. Biosyst. 2010, 6, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, A.; Wengel, J. Thermodynamics of RNA duplexes modified with unlocked nucleic acid nucleotides. Nucleic Acids Res. 2010, 38, 6697–6706. [Google Scholar] [CrossRef] [PubMed]
- Alagia, A.; Terrazas, M.; Eritja, R. RNA/aTNA chimeras: RNAi effects and nucleases resistance of single and double stranded RNAs. Molecules 2014, 19, 17872–17896. [Google Scholar] [CrossRef] [PubMed]
- Asanuma, H.; Toda, T.; Murayama, K.; Liang, X.; Kashida, H. Unexpectedly stable artificial duplex from flexible acyclic threoninol. J. Am. Chem. Soc. 2010, 132, 14702–14703. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, Y.; Takai, J.; Ito, H.; Murayama, K.; Kashida, H.; Asanuma, H. Enhancement of stability and activity of siRNA by terminal substitution with serinol nucleic acid (SNA). ChemBioChem 2014, 15, 2549–2555. [Google Scholar] [CrossRef] [PubMed]
- Kashida, H.; Murayama, K.; Toda, T.; Asanuma, H. Control of the chirality and helicity of oligomers of serinol nucleic acid (SNA) by sequence design. Angew. Chem. Int. Ed. Engl. 2011, 50, 1285–1288. [Google Scholar] [CrossRef] [PubMed]
- Snead, N.M.; Escamilla-Powers, J.R.; Rossi, J.J.; McCaffrey, A.P. 5' Unlocked nucleic acid modification improves siRNA targeting. Mol. Ther. Nucleic Acids 2013, 2, e103. [Google Scholar] [CrossRef] [PubMed]
- Haringsma, H.J.; Li, J.J.; Soriano, F.; Kenski, D.M.; Flanagan, W.M.; Willingham, A.T. MRNA knockdown by single strand RNA is improved by chemical modifications. Nucleic Acids Res. 2012, 40, 4125–4136. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, M.; Kitade, Y. Computational analysis of siRNA recognition by the Ago2 Paz domain and identification of the determinants of RNA-induced gene silencing. PLoS ONE 2013, 8, e57140. [Google Scholar] [CrossRef] [PubMed]
- Deleavey, G.F.; Frank, F.; Hassler, M.; Wisnovsky, S.; Nagar, B.; Damha, M.J. The 5' binding Mid domain of human Argonaute2 tolerates chemically modified nucleotide analogues. Nucleic Acid Ther. 2013, 23, 81–87. [Google Scholar] [PubMed]
- Jackson, A.L.; Burchard, J.; Leake, D.; Reynolds, A.; Schelter, J.; Guo, J.; Johnson, J.M.; Lim, L.; Karpilow, J.; Nichols, K.; et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 2006, 12, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Somoza, A.; Terrazas, M.; Eritja, R. Modified siRNAs for the study of the Paz domain. Chem. Commun. 2010, 46, 4270–4272. [Google Scholar] [CrossRef] [Green Version]
- Efthymiou, T.C.; Peel, B.; Huynh, V.; Desaulniers, J.P. Evaluation of siRNAs that contain internal variable-length spacer linkages. Bioorganic Med. Chem. Lett. 2012, 22, 5590–5594. [Google Scholar] [CrossRef]
- Valdmanis, P.N.; Gu, S.; Schuermann, N.; Sethupathy, P.; Grimm, D.; Kay, M.A. Expression determinants of mammalian Argonaute proteins in mediating gene silencing. Nucleic Acids Res. 2012, 40, 3704–3713. [Google Scholar] [CrossRef] [PubMed]
- Aleman, L.M.; Doench, J.; Sharp, P.A. Comparison of siRNA-induced off-target RNA and protein effects. RNA 2007, 13, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Caffrey, D.R.; Zhao, J.; Song, Z.; Schaffer, M.E.; Haney, S.A.; Subramanian, R.R.; Seymour, A.B.; Hughes, J.D. SiRNA off-target effects can be reduced at concentrations that match their individual potency. PLoS ONE 2011, 6, e21503. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Ui-Tei, K.; Naito, Y.; Nishi, K.; Juni, A.; Saigo, K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res. 2008, 36, 7100–7109. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Zhang, Y.; Jin, L.; Huang, Y.; Zhang, F.; Bassik, M.C.; Kampmann, M.; Kay, M.A. Weak base pairing in both seed and 3' regions reduces RNAi off-targets and enhances si/shRNA designs. Nucleic Acids Res. 2014, 42, 12169–12176. [Google Scholar] [CrossRef] [PubMed]
- Varani, G.; McClain, W.H. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000, 1, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Addepalli, H.; Meena; Peng, C.G.; Wang, G.; Fan, Y.; Charisse, K.; Jayaprakash, K.N.; Rajeev, K.G.; Pandey, R.K.; Lavine, G.; et al. Modulation of thermal stability can enhance the potency of siRNA. Nucleic Acids Res. 2010, 38, 7320–7331. [Google Scholar] [CrossRef] [PubMed]
- Bramsen, J.B.; Pakula, M.M.; Hansen, T.B.; Bus, C.; Langkjaer, N.; Odadzic, D.; Smicius, R.; Wengel, S.L.; Chattopadhyaya, J.; Engels, J.W.; et al. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010, 38, 5761–5773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrova, N.S.; Meschaninova, M.I.; Venyaminova, A.G.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Silencing activity of 2'-o-methyl modified anti-mdr1 siRNAs with mismatches in the central part of the duplexes. FEBS Lett. 2011, 585, 2352–2356. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ma, H.; Ye, C.; Ramirez, D.; Chen, S.; Montoya, J.; Shankar, P.; Wang, X.A.; Manjunath, N. Improved siRNA/shRNA functionality by mismatched duplex. PLoS ONE 2011, 6, e28580. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Jonsson, Z.O.; Dutta, A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 2003, 278, 44312–44319. [Google Scholar] [CrossRef] [PubMed]
- Holen, T.; Moe, S.E.; Sorbo, J.G.; Meza, T.J.; Ottersen, O.P.; Klungland, A. Tolerated wobble mutations in siRNAs decrease specificity, but can enhance activity in vivo. Nucleic Acids Res. 2005, 33, 4704–4710. [Google Scholar] [CrossRef] [PubMed]
- Leuschner, P.J.; Ameres, S.L.; Kueng, S.; Martinez, J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006, 7, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Rana, T.M. Illuminating the silence: Understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Juranek, S.; Li, H.; Sheng, G.; Tuschl, T.; Patel, D.J. Structure of an Argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 2008, 456, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Jin, L.; Zhang, F.; Huang, Y.; Grimm, D.; Rossi, J.J.; Kay, M.A. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc. Natl. Acad. Sci. USA 2011, 108, 9208–9213. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Liu, N.; Lai, E.C. Distinct mechanisms for microRNA strand selection by drosophila Argonautes. Mol. Cell 2009, 36, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Noland, C.L.; Doudna, J.A. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA 2013, 19, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Thonberg, H.; Wang, J.; Wahlestedt, C.; Liang, Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005, 33, 1671–1677. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.X.; Yang, J.; Sun, J.F.; Jia, L.T.; Zhang, Y.; Zhang, H.Z.; Li, X.; Meng, Y.L.; Yao, L.B.; Yang, A.G. Both strands of siRNA have potential to guide posttranscriptional gene silencing in mammalian cells. PLoS ONE 2009, 4, e5382. [Google Scholar] [CrossRef] [PubMed]
- O’Carroll, D.; Mecklenbrauker, I.; Das, P.P.; Santana, A.; Koenig, U.; Enright, A.J.; Miska, E.A.; Tarakhovsky, A. A slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007, 21, 1999–2004. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-blast: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are not available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alagia, A.; Terrazas, M.; Eritja, R. Modulation of the RNA Interference Activity Using Central Mismatched siRNAs and Acyclic Threoninol Nucleic Acids (aTNA) Units. Molecules 2015, 20, 7602-7619. https://doi.org/10.3390/molecules20057602
Alagia A, Terrazas M, Eritja R. Modulation of the RNA Interference Activity Using Central Mismatched siRNAs and Acyclic Threoninol Nucleic Acids (aTNA) Units. Molecules. 2015; 20(5):7602-7619. https://doi.org/10.3390/molecules20057602
Chicago/Turabian StyleAlagia, Adele, Montserrat Terrazas, and Ramon Eritja. 2015. "Modulation of the RNA Interference Activity Using Central Mismatched siRNAs and Acyclic Threoninol Nucleic Acids (aTNA) Units" Molecules 20, no. 5: 7602-7619. https://doi.org/10.3390/molecules20057602