Nkrp1 Family, from Lectins to Protein Interacting Molecules
Abstract
:1. Introduction
2. Nkrp1 Family
3. Structure of Nkrp1 Receptors
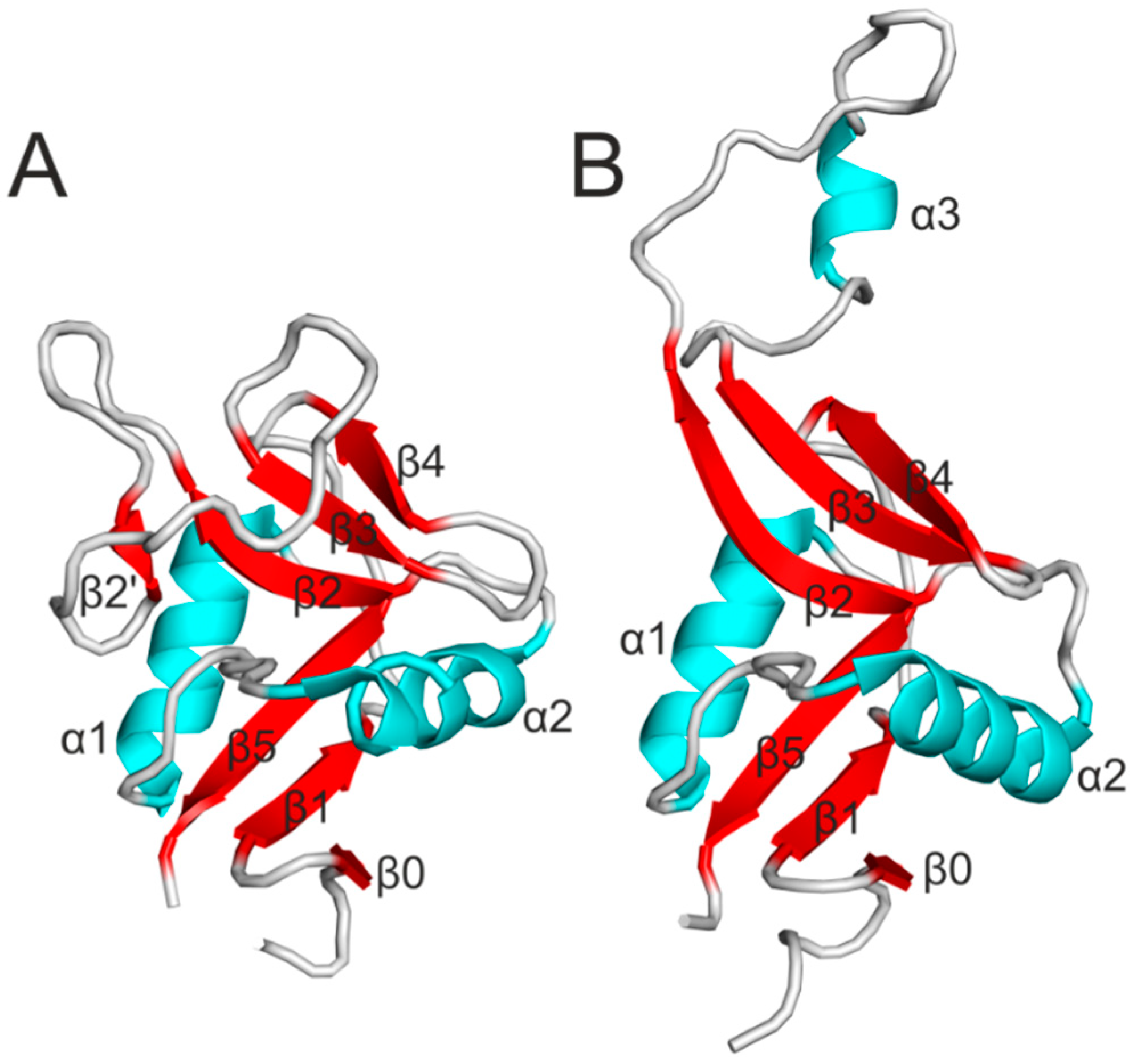
4. Signaling
5. Saccharide Ligands of Nkrp1—20 Years of Controversy
6. The Clr Ligands

7. Biological Relevance of Nkrp1:Clr Interactions
8. Conclusions
Acknowledgments
Author Contributions
Abbreviation
| CTLD | C-type lectin-like domain |
| GalNAc | 2-acetamido-2-deoxy-d-galactopyranose |
| GlcNAc | 2-acetamido-2-deoxy-d-glucopyranose |
| ITAM | immunoreceptor tyrosine-based activation motif |
| ITIM | immunoreceptor tyrosine-based inhibitory motif |
| LacdiNAc | 2-acetamido-2-deoxy-β-d-galactopyranosyl-(1→4)-2-acetamido-2-deoxy-d-glucopyranose |
| ManNAc | 2-acetamido-d-mannopyranose |
| NKC | NK cell gene complex |
| PAMAM dendrimer | polyamidoamine dendrimer |
Conflicts of Interest
References
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immunol. 2008, 9, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Strowig, T.; Brilot, F.; Münz, C. Noncytotoxic functions of NK cells: Direct pathogen restriction and assistance to adaptive immunity. J. Immunol. 2008, 180, 7785–7791. [Google Scholar] [CrossRef] [PubMed]
- Villard, J. The role of natural killer cells in human solid organ and tissue transplantation. J. Innate Immun. 2011, 3, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Gershwin, M.E.; Zhang, C. Regulatory NK cells in autoimmune disease. J. Autoimmun. 2012, 39, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Mathias, C.B.; Guernsey, L.A.; Zammit, D.; Brammer, C.; Wu, C.A.; Thrall, R.S.; Aguila, H.L. Pro-inflammatory role of natural killer cells in the development of allergic airway disease. Clin. Exp. Allergy 2014, 44, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Winger, E.E.; Reed, J.L. The multiple faces of the decidual natural killer cell. Am. J. Reprod. Immunol. 2013, 70, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L. NK cell recognition. Annu. Rev. Immunol. 2005, 23, 225–274. [Google Scholar] [CrossRef] [PubMed]
- Kärre, K.; Ljunggren, H.G.; Piontek, G.; Kiessling, R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 1986, 319, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Raulet, D.H.; Vance, R.E. Self-tolerance of natural killer cells. Nat. Rev. Immunol. 2006, 6, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Groh, V.; Wu, J.; Steinle, A.; Phillips, J.H.; Lanier, L.L.; Spies, T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 1999, 285, 727–729. [Google Scholar] [CrossRef] [PubMed]
- Giorda, R.; Rudert, W.; Vavassori, C.; Chambers, W.H.; Hiserodt, J.C.; Trucco, M. NKR-P1, a signal transduction molecule on natural killer cells. Science 1990, 249, 1298–300. [Google Scholar] [CrossRef] [PubMed]
- Giorda, R.; Trucco, M. Mouse NKR-P1. A family of genes selectively coexpressed in adherent lymphokine-activated killer cells. J. Immunol. 1991, 147, 1701–1708. [Google Scholar] [PubMed]
- Glimcher, L.; Shen, F.W.; Cantors, H. Identification of a cell-surface expressed antigen selectively on the natural killer cell. J. Exp. Med. 1977, 145, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ryan, J.C.; Turck, J.; Niemi, E.C.; Yokoyama, W.M.; Seaman, W.E. Molecular cloning of the NK1.1 antigen, a member of the NKR-P1 family of natural killer cell activation molecules. J. Immunol. 1992, 149, 1631–1635. [Google Scholar] [PubMed]
- Lanier, L.L.; Chang, C.; Phillips, J.H. Human NKR-P1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J. Immunol. 1994, 153, 2417–2428. [Google Scholar] [PubMed]
- Plougastel, B.; Matsumoto, K.; Dubbelde, C.; Yokoyama, W.M. Analysis of a 1-Mb BAC contig overlapping the mouse Nkrp1 cluster of genes: Cloning of three new Nkrp1 members, Nkrp1d, Nkrp1e, and Nkrp1f. Immunogenetics 2001, 53, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Kveberg, L.; Dai, K.-Z.; Westgaard, I.H.; Daws, M.R.; Fossum, S.; Naper, C.; Vaage, J.T. Two major groups of rat NKR-P1 receptors can be distinguished based on chromosomal localization, phylogenetic analysis and Clr ligand binding. Eur. J. Immunol. 2009, 39, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Kveberg, L.; Dai, K.-Z.; Inngjerdingen, M.; Brooks, C.G.; Fossum, S.; Vaage, J.T. Phylogenetic and functional conservation of the NKR-P1F and NKR-P1G receptors in rat and mouse. Immunogenetics 2011, 63, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, W.M.; Ryan, J.C.; Hunter, J.J.; Smith, H.R.C.; Stark, M.; Seamant, W.E. cDNA cloning of mouse NKR-P1 and genetic linkage with LY-49. Identification of a natural killer cell gene complex on chromosome 6. J. Immunol. 1991, 147, 3229–3236. [Google Scholar] [PubMed]
- Dissen, E.; Ryan, J.C.; Seaman, W.E.; Fossum, S. An Autosomal Dominant Locus, Nka, Mapping to the Ly-49 Region of a Rat Natural Killer (NK) Gene Complex, Controls NK Cell Lysis of Allogeneic Lymphocytes. J. Exp. Med. 1996, 183, 2197–2207. [Google Scholar] [CrossRef] [PubMed]
- Chambers, W.H.; Vujanovic, N.L.; DeLeo, A.B.; Olszowy, M.W.; Herberman, R.B.; Hiserodt, J.C. Monoclonal antibody to a triggering structure expressed on rat natural killer cells and adherent lymphokine-activated killer cells. J. Exp. Med. 1989, 169, 1373–1389. [Google Scholar] [CrossRef] [PubMed]
- Ryan, J.C.; Niemi, E.C.; Goldfien, R.D.; Hiserodt, J.C.; Seaman, W.E. NKR-P1, an activating molecule on rat natural killer cells, stimulates phosphoinositide turnover and a rise in intracellular calcium. J. Immunol. 1991, 3244–3250. [Google Scholar]
- Kveberg, L.; Bäck, C.J.; Dai, K.-Z.; Inngjerdingen, M.; Rolstad, B.; Ryan, J.C.; Vaage, J.T.; Naper, C. The novel inhibitory NKR-P1C receptor and Ly49s3 identify two complementary, functionally distinct NK cell subsets in rats. J. Immunol. 2006, 176, 4133–4140. [Google Scholar] [CrossRef] [PubMed]
- Appasamy, P.M.; Kenniston, T.W.; Brissette-Storkus, C.S.; Chambers, W.H. NKR-P1dim/TCR alpha beta + T cells and natural killer cells share expression of NKR-P1A and NKR-P1D. Nat. Immun. 1996, 15, 259–268. [Google Scholar] [PubMed]
- Aust, J.G.; Gays, F.; Mickiewicz, K.M.; Buchanan, E.; Brooks, C.G. The expression and function of the NKRP1 receptor family in C57BL/6 mice. J. Immunol. 2009, 183, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, K.; Naidenko, O.V.; Plougastel, B.F.M.; Fremont, D.H.; Yokoyama, W.M. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat. Immunol. 2003, 4, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Rozbeský, D.; Kavan, D.; Chmelík, J.; Novák, P.; Vaněk, O.; Bezouška, K. High-level expression of soluble form of mouse natural killer cell receptor NKR-P1C(B6) in Escherichia coli. Protein Expr. Purif. 2011, 77, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Weis, W.I.; Kahn, R.; Fourme, R.; Drickamer, K.; Hendrickson, W.A. Structure of the calcium-dependent lectin domain from a rat mannose-binding protein determined by MAD phasing. Science 1991, 254, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Zelensky, A.N.; Gready, J.E. The C-type lectin-like domain superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef] [PubMed]
- Sovová, Z.; Kopecký, V.; Pazderka, T.; Hofbauerová, K.; Rozbeský, D.; Vaněk, O.; Bezouška, K.; Ettrich, R. Structural analysis of natural killer cell receptor protein 1 (NKR-P1) extracellular domains suggests a conserved long loop region involved in ligand specificity. J. Mol. Model. 2011, 17, 1353–1370. [Google Scholar] [CrossRef] [PubMed]
- Kolenko, P.; Rozbeský, D.; Vaněk, O.; Kopecký, V.; Hofbauerová, K.; Novák, P.; Pompach, P.; Hašek, J.; Skálová, T.; Bezouška, K.; et al. Molecular architecture of mouse activating NKR-P1 receptors. J. Struct. Biol. 2011, 175, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, H.; Park-Snyder, S.; Kolatkar, R.; Heise, C.T.; Taylor, M.E.; Weis, W.I. Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor. J. Biol. Chem. 2000, 275, 21539–21548. [Google Scholar] [CrossRef] [PubMed]
- Maita, N.; Nishio, K.; Nishimoto, E.; Matsui, T.; Shikamoto, Y.; Morita, T.; Sadler, J.E.; Mizuno, H. Crystal structure of von Willebrand factor A1 domain complexed with snake venom, bitiscetin: Insight into glycoprotein Ib alpha binding mechanism induced by snake venom proteins. J. Biol. Chem. 2003, 278, 37777–37781. [Google Scholar] [CrossRef] [PubMed]
- Rozbesky, D.; Man, P.; Kavan, D.; Chmelik, J.; Cerny, J.; Bezouska, K.; Novak, P. Chemical cross-linking and H/D exchange for fast refinement of protein crystal structure. Anal. Chem. 2012, 84, 867–870. [Google Scholar] [CrossRef] [PubMed]
- Rozbesky, D.; Sovova, Z.; Marcoux, J.; Man, P.; Ettrich, R.; Robinson, C.V.; Novak, P. Structural model of lymphocyte receptor NKR-P1C revealed by mass spectrometry and molecular modeling. Anal. Chem. 2013, 85, 1597–1604. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Q.; Chen, S.; Brown, P.H.; Mariuzza, R.A. Structure of NKp65 bound to its keratinocyte ligand reveals basis for genetically linked recognition in natural killer gene complex. Proc. Natl. Acad. Sci. USA 2013, 110, 11505–11510. [Google Scholar] [CrossRef] [PubMed]
- Carlyle, J.R.; Martin, A.; Mehra, A.; Attisano, L.; Tsui, F.W.; Zúñiga-Pflücker, J.C. Mouse NKR-P1B, a novel NK1.1 antigen with inhibitory function. J. Immunol. 1999, 162, 5917–5923. [Google Scholar] [PubMed]
- Li, J.; Rabinovich, B.A.; Hurren, R.; Shannon, J.; Miller, R.G. Expression cloning and function of the rat NK activating and inhibitory receptors NKR-P1A and -P1B. Int. Immunol. 2003, 15, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L. Up on the tightrope: Natural killer cell activation and inhibition. Nat. Immunol. 2008, 9, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Ljutic, B.; Carlyle, J.R.; Filipp, D.; Nakagawa, R.; Julius, M.; Zúñiga-Pflücker, J.C. Functional requirements for signaling through the stimulatory and inhibitory mouse NKR-P1 (CD161) NK cell receptors. J. Immunol. 2005, 174, 4789–4796. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Bélanger, S.; Aguilar, O.; Zhang, Q.; St-Laurent, A.; Rahim, M.M.A.; Makrigiannis, A.P.; Carlyle, J.R. Analysis of the mouse 129-strain Nkrp1-Clr gene cluster reveals conservation of genomic organization and functional receptor-ligand interactions despite significant allelic polymorphism. Immunogenetics 2011, 63, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E.; Nunès, J.; Vély, F. Natural killer cell signaling pathways. Science 2004, 306, 1517–1519. [Google Scholar] [CrossRef] [PubMed]
- Burshtyn, D.N.; Yang, W.; Yi, T.; Long, E.O. A novel phosphotyrosine motif with a critical amino acid at position -2 for the SH2 domain-mediated activation of the tyrosine phosphatase SHP-1. J. Biol. Chem. 1997, 272, 13066–13072. [Google Scholar] [CrossRef] [PubMed]
- Tamir, I.; dal Porto, J.M.; Cambier, J.C. Cytoplasmic protein tyrosine phosphatases SHP-1 and SHP-2: Regulators of B cell signal transduction. Curr. Opin. Immunol. 2000, 12, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Ravetch, J.V.; Lanier, L.L. Immune Inhibitory Receptors. Science 2000, 290, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Pozo, D.; Valés-Gómez, M.; Mavaddat, N.; Williamson, S.C.; Chisholm, S.E.; Reyburn, H. CD161 (human NKR-P1A) signaling in NK cells involves the activation of acid sphingomyelinase. J. Immunol. 2006, 176, 2397–2406. [Google Scholar] [CrossRef] [PubMed]
- Bezouska, K.; Vlahas, G.; Horváth, O.; Jinochová, G.; Fiserová, A.; Giorda, R.; Chambers, W.H.; Feizi, T.; Pospísil, M. Rat natural killer cell antigen, NKR-P1, related to C-type animal lectins is a carbohydrate-binding protein. J. Biol. Chem. 1994, 269, 16945–16952. [Google Scholar] [PubMed]
- Bezouska, K.; Yuen, C.T.; O’Brien, J.; Childs, R.A.; Chai, W.; Lawson, A.M.; Drbal, K.; Fiserová, A.; Pospísil, M.; Feizi, T. Oligosaccharide ligands for NKR-P1 protein activate NK cells and cytotoxicity. Nature 1994, 372, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Bezouska, K.; Yuen, C.T.; O’Brien, J.; Childs, R.A.; Chai, W.; Lawson, A.M.; Drbal, K.; Fiserová, A.; Pospisil, M.; Feizi, T. Correspondence: Oligosaccharide ligands for NKR-P1 protein activate NK cells and cytotoxicity. Nature 1996, 380, 559–559. [Google Scholar]
- Kogelberg, H.; Montero, E.; Bay, S.; Lawson, A.M.; Feizi, T. Re-evaluation of monosaccharide binding property of recombinant soluble carbohydrate recognition domain of the natural killer cell receptor NKR-P1A. J. Biol. Chem. 1999, 274, 30335–30336. [Google Scholar] [PubMed]
- Bezouska, K.; Sklenar, J.; Dvorakova, J.; Havlicek, V.; Pospisil, M.; Thiem, J.; Kren, V. NKR-P1A protein, an activating receptor of rat natural killer cells, binds to the chitobiose core ofuncompletely glycosylated N-linked glycans, and to linear chitooligomers. Biochem. Biophys. Res. Commun. 1997, 238, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Krist, P.; Herkommerová-Rajnochová, E.; Rauvolfová, J.; Semenuk, T.; Vavrusková, P.; Pavlícek, J.; Bezouska, K.; Petrus, L.; Kren, V. Toward an optimal oligosaccharide ligand for rat natural killer cell activation receptor NKR-P1. Biochem. Biophys. Res. Commun. 2001, 287, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Sedmera, P.; Přikrylová, V.; Bezouška, K.; Rajnochová, E.; Thiem, J.; Křen, V. Preparation of mannac containing chitooligomers by isomerisation and their binding to Nkr-P1 Protein. J. Carbohydr. Chem. 1998, 17, 1351–1357. [Google Scholar] [CrossRef]
- Kren, V.; Dvoráková, J.; Gambert, U.; Sedmera, P.; Havlícek, V.; Thiem, J.; Bezouska, K. beta-Glucosylation of chitooligomers by galactosyltransferase. Carbohydr. Res. 1997, 305, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Semenuk, T.; Krist, P.; Pavlícek, J.; Bezouska, K.; Kuzma, M.; Novák, P.; Kren, V. Synthesis of chitooligomer-based glycoconjugates and their binding to the rat natural killer cell activation receptor NKR-P1. Glycoconj. J. 2001, 18, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Bezouska, K.; Kren, V.; Kieburg, C.; Lindhorst, T.K. GlcNAc-terminated glycodendrimers form defined precipitates with the soluble dimeric receptor of rat natural killer cells, sNKR-P1A. FEBS Lett. 1998, 426, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Pospisil, M.; Vannucci, L.; Fiserova, A.; Krausova, K.; Horvath, O.; Kren, V.; Mosca, F.; Lindhorst, T.K.; Sadalapure, K.; Bezouska, K. Glycodendrimeric ligands of c-type lectin receptors as therapeutic agents in experimental cancer. Adv. Exp. Med. Biol. 2001, 495, 343–347. [Google Scholar] [PubMed]
- Krist, P.; Vannucci, L.; Kuzma, M.; Man, P.; Sadalapure, K.; Patel, A.; Bezouska, K.; Pospísil, M.; Petrus, L.; Lindhorst, T.K.; Kren, V. Fluorescent labelled thiourea-bridged glycodendrons. Chembiochem 2004, 5, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Vannucci, L.; Fiserová, A.; Sadalapure, K.; Lindhorst, T.K.; Kuldová, M.; Rossmann, P.; Horváth, O.; Kren, V.; Krist, P.; Bezouska, K.; et al. Effects of N-acetyl-glucosamine-coated glycodendrimers as biological modulators in the B16F10 melanoma model in vivo. Int. J. Oncol. 2003, 23, 285–296. [Google Scholar] [PubMed]
- Attolino, E.; Bonaccorsi, F.; Catelani, G.; D’Andrea, F.; Křenek, K.; Bezouška, K.; Křen, V. Improved preparation of β-d-ManNAc-(1→4)-D-Glc and β-d-TalNAc-(1→4)-d-Glc disaccharides and evaluation of their activating properties on the Natural Killer cells NKR-P1 and CD69 Receptors. J. Carbohydr. Chem. 2008, 27, 156–171. [Google Scholar] [CrossRef]
- Bojarová, P.; Krenek, K.; Wetjen, K.; Adamiak, K.; Pelantová, H.; Bezouska, K.; Elling, L.; Kren, V. Synthesis of LacdiNAc-terminated glycoconjugates by mutant galactosyltransferase-a way to new glycodrugs and materials. Glycobiology 2009, 19, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Drozdová, A.; Bojarová, P.; Křenek, K.; Weignerová, L.; Henssen, B.; Elling, L.; Christensen, H.; Jensen, H.H.; Pelantová, H.; Kuzma, M.; et al. Enzymatic synthesis of dimeric glycomimetic ligands of NK cell activation receptors. Carbohydr. Res. 2011, 346, 1599–1609. [Google Scholar] [CrossRef] [PubMed]
- Bojarová, P.; Křenek, K.; Kuzma, M.; Petrásková, L.; Bezouška, K.; Namdjou, D.-J.; Elling, L.; Křen, V. N-Acetylhexosamine triad in one molecule: Chemoenzymatic introduction of 2-acetamido-2-deoxy-β-d-galactopyranosyluronic acid residue into a complex oligosaccharide. J. Mol. Catal. B Enzym. 2008, 50, 69–73. [Google Scholar] [CrossRef]
- Fialová, P.; Namdjou, D.-J.; Ettrich, R.; Přikrylová, V.; Rauvolfová, J.; Křenek, K.; Kuzma, M.; Elling, L.; Bezouška, K.; Křen, V. Combined application of galactose oxidase and β-N-acetylhexosaminidase in the synthesis of complex immunoactive N-acetyl-d-galactosaminides. Adv. Synth. Catal. 2005, 347, 997–1006. [Google Scholar] [CrossRef]
- Bojarová, P.; Slámová, K.; Křenek, K.; Gažák, R.; Kulik, N.; Ettrich, R.; Pelantová, H.; Kuzma, M.; Riva, S.; Adámek, D.; et al. Charged hexosaminides as new substrates for β-N-acetylhexosaminidase-catalyzed synthesis of immunomodulatory disaccharides. Adv. Synth. Catal. 2011, 353, 2409–2420. [Google Scholar] [CrossRef]
- Catelani, G.; D’Andrea, F.; Griselli, A.; Guazzelli, L.; Nemcová, P.; Bezouska, K.; Krenek, K.; Kren, V. Deoxynojirimycin and its hexosaminyl derivatives bind to natural killer cell receptors rNKR-P1A and hCD69. Bioorg. Med. Chem. Lett. 2010, 20, 4645–4648. [Google Scholar] [CrossRef] [PubMed]
- Slámová, K.; Marhol, P.; Bezouska, K.; Lindkvist, L.; Hansen, S.G.; Kren, V.; Jensen, H.H. Synthesis and biological activity of glycosyl-1H-1,2,3-triazoles. Bioorg. Med. Chem. Lett. 2010, 20, 4263–4265. [Google Scholar] [CrossRef] [PubMed]
- Krenek, K.; Kuldová, M.; Hulíková, K.; Stibor, I.; Lhoták, P.; Dudic, M.; Budka, J.; Pelantová, H.; Bezouska, K.; Fiserová, A.; et al. N-acetyl-d-glucosamine substituted calix[4]arenes as stimulators of NK cell-mediated antitumor immune response. Carbohydr. Res. 2007, 342, 1781–1792. [Google Scholar] [CrossRef] [PubMed]
- Hulikova, K.; Benson, V.; Svoboda, J.; Sima, P.; Fiserova, A. N-Acetyl-d-glucosamine-coated polyamidoamine dendrimer modulates antibody formation via natural killer cell activation. Int. Immunopharmacol. 2009, 9, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Hulíková, K.; Grobárová, V.; Křivohlavá, R.; Fišerová, A. Antitumor activity of N-acetyl-d-glucosamine-substituted glycoconjugates and combined therapy with keyhole limpet hemocyanin in B16F10 mouse melanoma model. Folia Microbiol. (Praha). 2010, 55, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Hulikova, K.; Svoboda, J.; Benson, V.; Grobarova, V.; Fiserova, A. N-acetyl-d-glucosamine-coated polyamidoamine dendrimer promotes tumor-specific B cell responses via natural killer cell activation. Int. Immunopharmacol. 2011, 11, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Bezouska, K.; Nepovim, A.; Horvath, O.; Pospisil, M.; Hamann, J.; Feizi, T. CD69 antigen of human lymphocytes is a calcium-dependent carbohydrate-binding protein. Biochem. Biophys. Res. Commun. 1995, 208, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Pavlicek, J.; Sopko, B.; Ettrich, R.; Kopecky, V.; Baumruk, V.; Man, P.; Havlicek, V.; Vrbacky, M.; Martinkova, L.; Kren, V.; et al. Molecular characterization of binding of calcium and carbohydrates by an early activation antigen of lymphocytes CD69. Biochemistry 2003, 42, 9295–9306. [Google Scholar] [CrossRef] [PubMed]
- Kavan, D.; Kubickova, M.; Bily, J.; Vanek, O.; Hofbauerova, K.; Mrazek, H.; Rozbesky, D.; Bojarova, P.; Kren, V.; Zidek, L.; et al. Cooperation between subunits is essential for high-affinity binding of N-acetyl-d-hexosamines to dimeric soluble and dimeric cellular forms of human CD69. Biochemistry 2010, 49, 4060–4067. [Google Scholar] [CrossRef] [PubMed]
- Kovalova, A.; Ledvina, M.; Saman, D.; Zyka, D.; Kubickova, M.; Zidek, L.; Sklenar, V.; Pompach, P.; Kavan, D.; Bily, J.; et al. Synthetic N-acetyl-d-glucosamine based fully branched tetrasaccharide, a mimetic of the endogenous ligand for CD69, activates CD69+ killer lymphocytes upon dimerization via a hydrophilic flexible linker. J. Med. Chem. 2010, 53, 4050–4065. [Google Scholar] [CrossRef] [PubMed]
- Report of the Joint Ethical Committee of the Institute of Microbiology, Prague and Charles University in Prague. English translation can be found in the Supplementary Material. Available online: http://www.biomed.cas.cz/mbu/doc/VyjadreniEK.PDF (accessed on 11 November 2014).
- Rozbeský, D.; Krejzová, J.; Křenek, K.; Prchal, J.; Hrabal, R.; Kožíšek, M.; Weignerová, L.; Fiore, M.; Dumy, P.; Křen, V.; et al. Re-evaluation of binding properties of recombinant lymphocyte receptors NKR-P1A and CD69 to chemically synthesized glycans and peptides. Int. J. Mol. Sci. 2014, 15, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Grobárová, V.; Benson, V.; Rozbeský, D.; Novák, P.; Cerný, J. Re-evaluation of the involvement of NK cells and C-type lectin-like NK receptors in modulation of immune responses by multivalent GlcNAc-terminated oligosaccharides. Immunol. Lett. 2013, 156, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Daniels, B.F.; Nakamura, M.C.; Rosen, S.D.; Yokoyama, W.M.; Seaman, W.E. Ly-49A, a receptor for H-2Dd, has a functional carbohydrate recognition domain. Immunity 1994, 1, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, J.; Tran, T.-V.; Kaudeer, J.; Oberle, K.; Herrmann, J.; Quagliano, I.; Abel, T.; Cohnen, A.; Gatterdam, V.; Jacobs, A.; et al. The stalk domain and the glycosylation status of the activating natural killer cell receptor NKp30 are important for ligand binding. J. Biol. Chem. 2012, 287, 31527–31539. [Google Scholar] [CrossRef] [PubMed]
- Mason, L.H.; Willette-Brown, J.; Anderson, S.K.; Alvord, W.G.; Klabansky, R.L.; Young, H.A.; Ortaldo, J.R. Receptor Glycosylation Regulates Ly-49 Binding to MHC Class I. J. Immunol. 2003, 171, 4235–4242. [Google Scholar] [CrossRef] [PubMed]
- Andresen, L.; Skovbakke, S.L.; Persson, G.; Hagemann-Jensen, M.; Hansen, K.A.; Jensen, H.; Skov, S. 2-deoxy d-glucose prevents cell surface expression of NKG2D ligands through inhibition of N-linked glycosylation. J. Immunol. 2012, 188, 1847–1855. [Google Scholar] [CrossRef] [PubMed]
- Plougastel, B.; Dubbelde, C.; Yokoyama, W.M. Cloning of Clr, a new family of lectin-like genes localized between mouse Nkrp1a and Cd69. Immunogenetics 2001, 53, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Kartsogiannis, V.; Hu, Y.S.; Elliott, J.; Quinn, J.M.; McKinstry, W.J.; Gillespie, M.T.; Ng, K.W. A novel osteoblast-derived C-type lectin that inhibits osteoclast formation. J. Biol. Chem. 2001, 276, 14916–14923. [Google Scholar] [CrossRef] [PubMed]
- Carlyle, J.R.; Jamieson, A.M.; Gasser, S.; Clingan, C.S.; Arase, H.; Raulet, D.H. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc. Natl. Acad. Sci. USA 2004, 101, 3527–3532. [Google Scholar] [CrossRef] [PubMed]
- Kamishikiryo, J.; Fukuhara, H.; Okabe, Y.; Kuroki, K.; Maenaka, K. Molecular basis for LLT1 protein recognition by human CD161 protein (NKRP1A/KLRB1). J. Biol. Chem. 2011, 286, 23823–23830. [Google Scholar] [CrossRef] [PubMed]
- Skálová, T.; Kotýnková, K.; Dušková, J.; Hašek, J.; Koval, T.; Kolenko, P.; Novák, P.; Man, P.; Hanč, P.; Vaněk, O.; et al. Mouse Clr-g, a ligand for NK cell activation receptor NKR-P1F: Crystal structure and biophysical properties. J. Immunol. 2012, 189, 4881–4889. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.B.; Bettadapura, J.; Alsharifi, M.; Mathew, P.; Warren, H.S.; Lanier, L.L. Cutting edge: Lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J. Immunol. 2005, 175, 7796–7799. [Google Scholar] [CrossRef] [PubMed]
- Aldemir, H.; Prod’homme, V.; Dumaurier, M.-J.; Retiere, C.; Poupon, G.; Cazareth, J.; Bihl, F.; Braud, V.M. Cutting Edge: Lectin-Like Transcript 1 Is a Ligand for the CD161 Receptor. J. Immunol. 2005, 175, 7791–7795. [Google Scholar] [CrossRef] [PubMed]
- Borrego, F.; Ulbrecht, M.; Weiss, E.H.; Coligan, J.E.; Brooks, A.G. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J. Exp. Med. 1998, 187, 813–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Söderström, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Llano, M.; Carretero, M.; Ishitani, A.; Navarro, F.; López-Botet, M.; Geraghty, D.E. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA 1998, 95, 5199–5204. [Google Scholar] [CrossRef] [PubMed]
- Mistry, A.R.; O’Callaghan, C.A. Regulation of ligands for the activating receptor NKG2D. Immunology 2007, 121, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Rahim, M.M.; Allan, D.S.J.; Tu, M.M.; Belanger, S.; Abou-Samra, E.; Ma, J.; Sekhon, H.S.; Fairhead, T.; Zein, H.S.; et al. Mouse Nkrp1-Clr gene cluster sequence and expression analyses reveal conservation of tissue-specific MHC-independent immunosurveillance. PLoS One 2012, 7, e50561. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.J.N.; Wilson, E.; Davidson, C.L.; Aguilar, O.; Fu, L.; Carlyle, J.R.; Burshtyn, D.N. Poxvirus infection-associated downregulation of C-type lectin-related-b prevents NK cell inhibition by NK receptor protein-1B. J. Immunol. 2012, 188, 4980–4991. [Google Scholar] [CrossRef] [PubMed]
- Fine, J.H.; Chen, P.; Mesci, A.; Allan, D.S.J.; Gasser, S.; Raulet, D.H.; Carlyle, J.R. Chemotherapy-induced genotoxic stress promotes sensitivity to natural killer cell cytotoxicity by enabling missing-self recognition. Cancer Res. 2010, 70, 7102–7113. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Nunez, R.; Cheng, S.; Ding, Y.; Tumang, J.; Lyddane, C.; Roman, C.; Liou, H.-C. C-type lectin OCILRP2/Clr-g and its ligand NKRP1f costimulate T cell proliferation and IL-2 production. Cell Immunol. 2005, 234, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Voigt, S.; Mesci, A.; Ettinger, J.; Fine, J.H.; Chen, P.; Chou, W.; Carlyle, J.R. Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1B:Clr-b missing-self axis. Immunity 2007, 26, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Fodil-Cornu, N.; Lee, S.-H.; Belanger, S.; Makrigiannis, A.P.; Biron, C.A.; Buller, R.M.; Vidal, S.M. Ly49h-deficient C57BL/6 mice: A new mouse cytomegalovirus-susceptible model remains resistant to unrelated pathogens controlled by the NK gene complex. J. Immunol. 2008, 181, 6394–6405. [Google Scholar] [CrossRef] [PubMed]
- Germain, C.; Meier, A.; Jensen, T.; Knapnougel, P.; Poupon, G.; Lazzari, A.; Neisig, A.; Håkansson, K.; Dong, T.; Wagtmann, N.; et al. Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-γ contributes to modulate immune responses. J. Biol. Chem. 2011, 286, 37964–37975. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.B.; Cao, W.; Avery, D.T.; Tangye, S.G.; Liu, Y.-J.; Houchins, J.P.; Lanier, L.L. Functional consequences of interactions between human NKR-P1A and its ligand LLT1 expressed on activated dendritic cells and B cells. J. Immunol. 2008, 180, 6508–6517. [Google Scholar] [CrossRef] [PubMed]
- Satkunanathan, S.; Kumar, N.; Bajorek, M.; Purbhoo, M.; Culley, F.J. Respiratory syncytial virus infection, TLR3 ligands, and proinflammatory cytokines induce CD161 ligand LLT1 expression on the respiratory epithelium. J. Virol. 2014, 88, 2366–2373. [Google Scholar] [CrossRef] [PubMed]
- Kolenko, P.; Rozbeský, D.; Vaněk, O.; Bezouška, K.; Hašek, J.; Dohnálek, J. Structure of the H107R variant of the extracellular domain of mouse NKR-P1A at 2.3 Å resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rozbeský, D.; Ivanova, L.; Hernychová, L.; Grobárová, V.; Novák, P.; Černý, J. Nkrp1 Family, from Lectins to Protein Interacting Molecules. Molecules 2015, 20, 3463-3478. https://doi.org/10.3390/molecules20023463
Rozbeský D, Ivanova L, Hernychová L, Grobárová V, Novák P, Černý J. Nkrp1 Family, from Lectins to Protein Interacting Molecules. Molecules. 2015; 20(2):3463-3478. https://doi.org/10.3390/molecules20023463
Chicago/Turabian StyleRozbeský, Daniel, Ljubina Ivanova, Lucie Hernychová, Valéria Grobárová, Petr Novák, and Jan Černý. 2015. "Nkrp1 Family, from Lectins to Protein Interacting Molecules" Molecules 20, no. 2: 3463-3478. https://doi.org/10.3390/molecules20023463





