Salvianolic Acid Y: A New Protector of PC12 Cells against Hydrogen Peroxide-Induced Injury from Salvia officinalis
Abstract
:1. Introduction
| Position | TSL 1 | Salvianolic Acid B | ||
|---|---|---|---|---|
| δH (ppm) | δc (ppm) | δH (ppm) | δc (ppm) | |
| 2 | 5.90 (d,9.2) | 87.0 | 5.81 (d,4.8) | 86.7 |
| 3 | 4.77 (d,9.2) | 53.2 | 4.30 (d,4.8) | 56.4 |
| 4 | - | 123.2 | - | 123.1 |
| 5 | 7.11 (d, 8.5) | 121.3 | 7.11 (d, 8.5) | 120.6 |
| 6 | 6.77 (d, 8.4) | 116.9 | 6.78 (d, 8.4) | 116.7 |
| 7 | - | 145.4 | - | 145.1 |
| 8 | - | 148.2 | - | 147.5 |
| 9 | - | 126.8 | - | 123.1 |
| 1' | - | 127.8 | - | 127.7 |
| 2' | 6.90 (s) | 113.6 | 6.69 | 111.8 |
| 3' | - | 144.4 | - | 144.2 |
| 4' | - | 143.9 | - | 143.6 |
| 5' | 6.68 (ov) | 114.7 | 6.70 (ov) | 114.8 |
| 6' | 6.68 (ov) | 118.4 | 6.70 (ov | 116.8 |
| 1'' | - | 126.9 | - | 127.4 |
| 2'' | 6.48 (d,1.9) | 116.4 | 6.47 (d,1.9) | 115.7 |
| 3'' | - | 143.8 | - | 143.4 |
| 4'' | - | 143.7 | - | 143.4 |
| 5'' | 6.56 (d,8.0) | 114.9 | 6.49 (d,8.0) | 114.9 |
| 6'' | 6.33 (dd,8.1,1.9) | 120.7 | 6.26 (dd,8.0,1.9) | 120.1 |
| 7''α | 2.54 (dd,14.0,6.1) | 36.1 | 2.94 (ov) | 35.9 |
| 7''β | 2.45 (dd,14.0,6.6) | - | 2.78 (dd,16.0,8.0) | - |
| 8'' | 4.35 (t, 6.3) | 74.4 | 5.11 (t, 4.0) | 74.1 |
| 9'' | - | 171.0 | - | 171.1 |
| 10'' | - | 170.0 | - | 170.7 |
| 1''' | - | 127.7 | - | 127.7 |
| 2''' | 6.71 (d,1.9) | 116.4 | 6.70 (d,1.9) | 115.7 |
| 3''' | - | 144.8 | - | 145.0 |
| 4''' | - | 144.6 | - | 144.3 |
| 5''' | 6.64 (d, 8.0) | 114.9 | 6.65 (d, 8.0) | 114.8 |
| 6''' | 6.57 (dd, 8.0, 1.9) | 120.8 | 6.57 (dd, 8.0, 1.9) | 120.1 |
| 7'''α | 3.02 (dd,14.3,4.8) | 36.4 | 3.03 (dd,16.0,4.0) | 36.3 |
| 7'''β | 2.98 (dd,14.2,6.9) | - | 2.95 (ov) | - |
| 8''' | 5.11 (t, 6.7) | 73.3 | 5.11 (t, 4.0) | 73.2 |
| 9''' | - | 171.2 | - | 172.2 |
| 10''' | - | 166.7 | - | 166.4 |
| 11''' | 6. 25 (d,16.0) | 115.5 | 6. 15 (d,16.0) | 115.0 |
| 12''' | 7.53 (d, 16.0) | 142.4 | 7.46 (d, 16.0) | 141.9 |
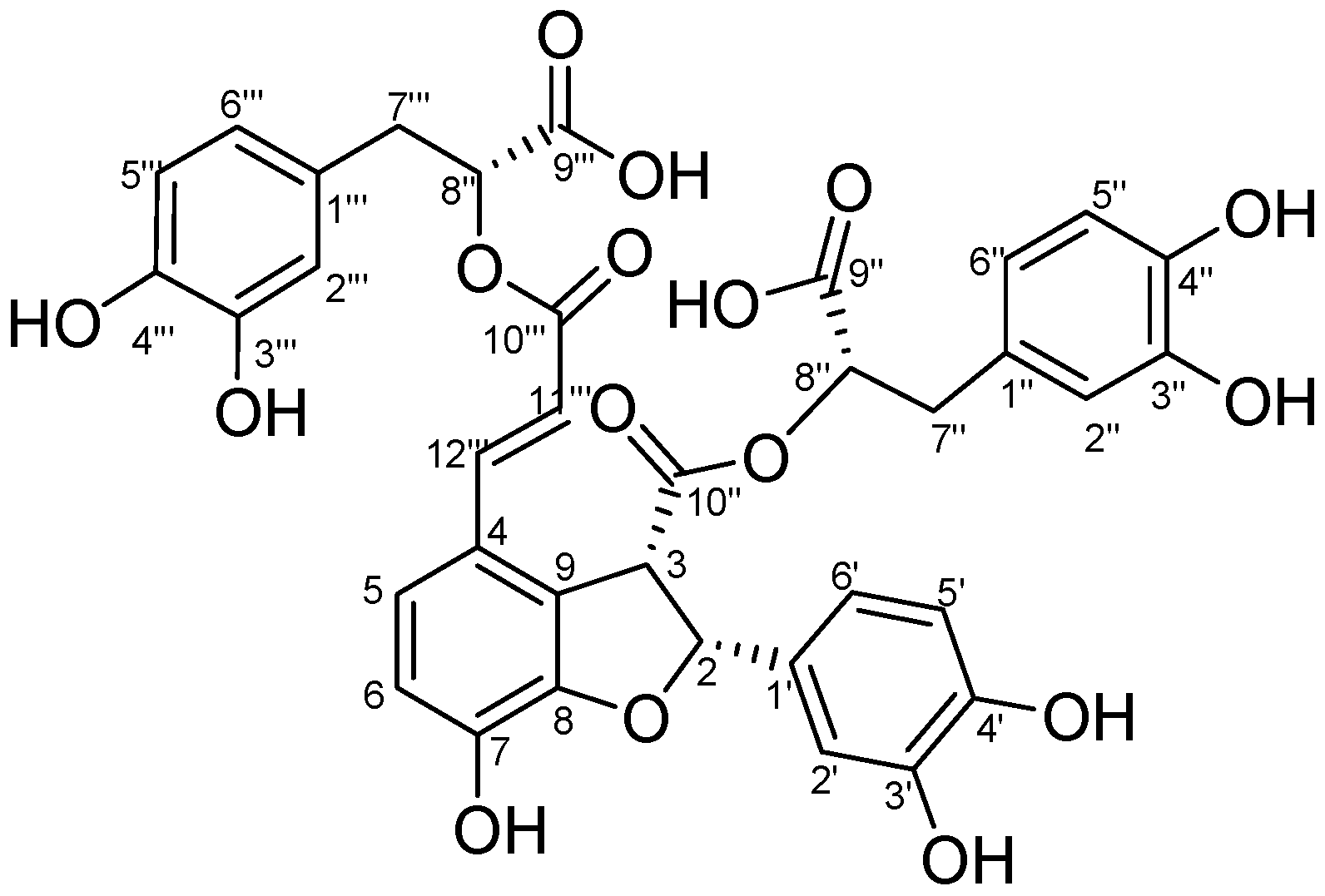
2. Results and Discussion
2.1. Structural Elucidation of Salvianolic Acid Y


2.2. In Vitro Biological Activity
| Group | Dose (μM) | Cell Viability (%) | Protection Rate (%) |
|---|---|---|---|
| control group | - | 100 ± 6.5 | - |
| model group | - | 49.3 ± 6.0 ## | - |
| salvianolic acid B | 10 | 68.7 ± 3.0 ** | 35.2 ± 6.2 |
| salvianolic acid Y | 10 | 79.2 ± 2.6 ** | 54.2 ± 5.4 @@ |
3. Experimental Section
3.1. General
3.2. Compound Isolation
3.3. Activity in Vitro
3.4. Statistical Analysis
3.5. Biosynthetic Pathway
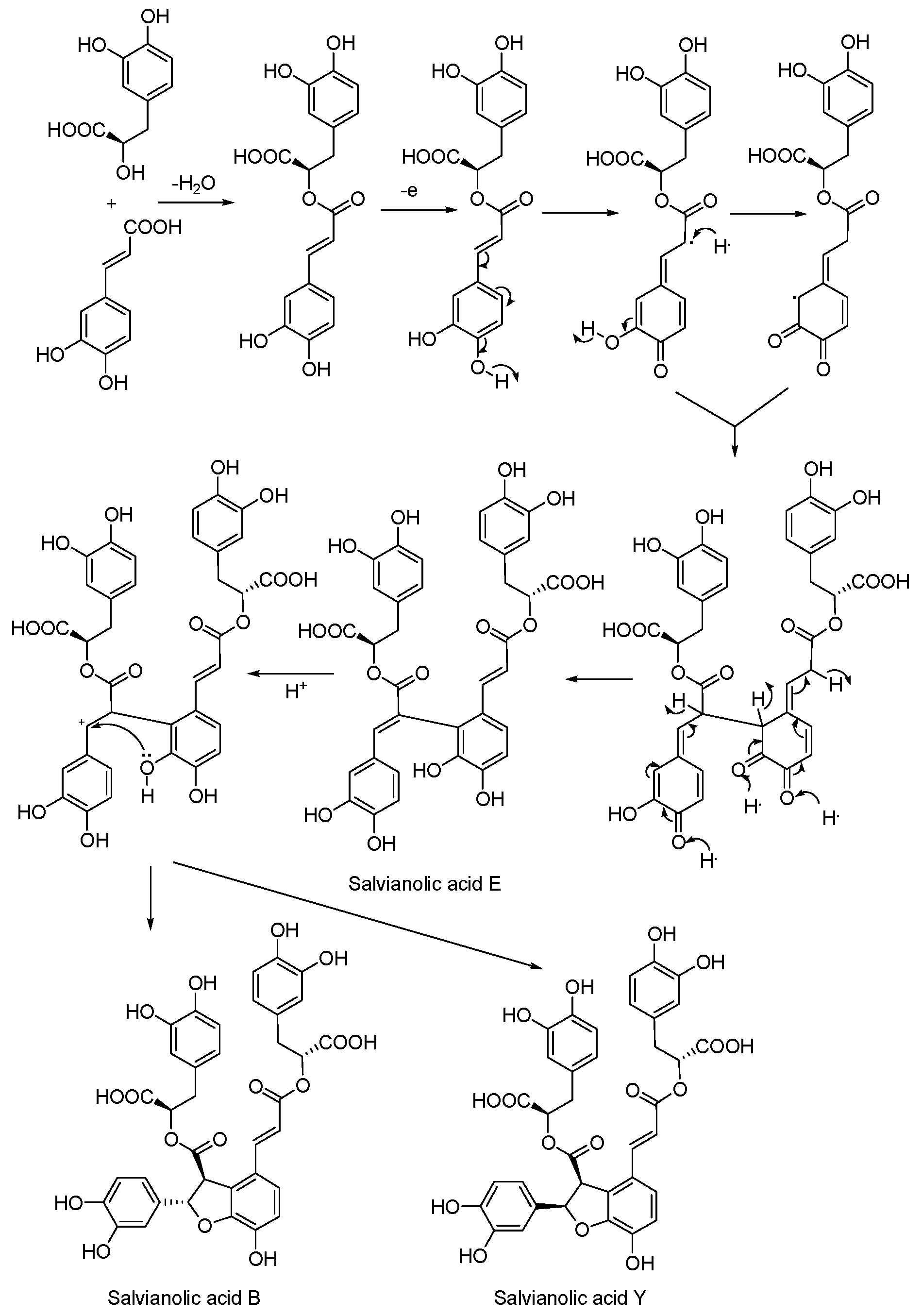
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lu, Y.R.; Foo, L.Y. Polyphenolics of Salvia—A review. Phytochemistry 2002, 59, 117–140. [Google Scholar] [CrossRef]
- Liu, J.; Dai, Z.; Wang, G.L.; Lin, R.C. Progress in bioactive constituents and isolation and analysis method of Salvia miltiorrhizae radix et ehizoma. Chin. J. Exp. Trad. Med. Form. 2012, 18, 288–295. [Google Scholar]
- Jiang, R.W.; Lau, K.; Hon, P.M.; Mark, T.C.W.; Woo, K.S.; Fung, K.P. Chemistry and biological activities of caffeic acid derivatives from Salvia miltiorrhiza. Curr. Med. Chem. 2005, 12, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.M.; Cheng, K.P.; Choang, T.F.; Chow, H.F.; Chui, K.Y.; Hon, P.M.; Tan, F.W.L.; Zhong, Z.P.; Lee, C.M.; Sham, H.L.; et al. Structure elucidation and total synthesis of new tanshinones isolated from Salvia miltiorrhizae Bunge (Danshen). J. Org. Chem. 1990, 55, 3537–3543. [Google Scholar] [CrossRef]
- Li, L.N. Water Soluble Active Components of Salvia miltiorrhiza and Related Plants. J. Chin. Pharm. Sci. 1997, 6, 57–64. [Google Scholar]
- Zan, Y.E.; Xu, J.P. Advances in pharmacological effects of salvianolic acid B. Mil. Med. J. S. Chin. 2007, 21, 37–39. [Google Scholar]
- Zhao, G.R.; Zhang, H.M.; Ye, T.X.; Xiang, Z.J.; Yuan, Y.J.; Guo, Z.X.; Zhao, L.B. Characterization of the radical scavenging and antioxidant activities of danshensu and salvianolic acid B. Food Chem. Toxicol. 2008, 46, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Gong, H.; Li, X.C.; Sun, Y.; Chen, H.; Zhang, X.; Wu, Q.; Zheng, L.; Huang, K. Salvianolic acid B inhibits the amyloid formation of human islet amyloid polypeptideand protects pancreatic β-cells against cytotoxicity. Proteins Struct. Funct. Bioinform. 2013, 81, 613–621. [Google Scholar] [CrossRef]
- Liu, M.; Ye, J.T.; Gao, S.; Fang, W.; Li, H.; Geng, B.; Zou, J.; Chen, X.; Chen, S.R.; Zhang, L.K.; et al. Salvianolic acid B protects cardiomyocytes from angiotensin II-induced hypertrophy via inhibition of PARP-1. Biochem. Biophys. Res. Commun. 2014, 444, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.H.; Xu, L.L; Zhou, C.H.; Wayne, Y.W. Lee; Wu, T.; Cui, L.; Li, G. Salvianolic acid B promotes osteogenesis of human mesenchymal stem cells through activating ERK signaling pathway. Int. J. Biochem. Cell Biol. 2014, 51, 1–9. [Google Scholar]
- Chen, Z.X.; Gu, W.H.; Huang, H.Z.; Yang, X.M.; Sun, C.J.; Chen, W.Z.; Dong, Y.L.; Ma, H.L. Studies of water-soluble phenolic acid components of Salvia. Chin. Pharm. Bull. 1981, 15, 24–25. [Google Scholar]
- Ai, C.B; Li, L.N. Stereostructure of salvianolic acid B and isolation of salvianolic acid C from Salvia miltiorrhiza. J. Nat. Prod. 1988, 51, 145–149. [Google Scholar] [CrossRef]
- Anja, W.; Steven, J.O.; Robert, G.B.; Jonathan, A.E. Reassignment of the configuration of salvianolic acid B and establishment of its identity with lithospermic acid B. J. Nat. Prod. 2006, 69, 1231–1233. [Google Scholar] [CrossRef] [PubMed]
- Wada, H.; Kido, T.; Tanaka, N.; Murakami, T.; Saiki, Y.; Chen, C.M. Chemical and chemotaxonomical studies of ferns. LXXXI. Characteristic lignans of Blechnaceous ferns. Chem. Pharm. Bull. 1992, 40, 2099–2101. [Google Scholar] [CrossRef]
- Zhang, Z.F.; Peng, Z.G.; Gao, L.; Dong, B.; Li, J.R.; Li, Z.Y.; Chen, H.S. Three new derivatives of anti-HIV-1 polyphenols isolated from Salvia yunnanensis. J. Asian Nat. Prod. Res. 2008, 10, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Nina, B.; Lorenzo, D.B.; Gennaro, P. Application of electronic circular dichroism in configurational and conformational analysis of organic compounds. Chem. Soc. Rev. 2007, 36, 914–931. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Mark, T.H. Atkamine: A New Pyrroloiminoquinone Scaffold from the Cold Water Aleutian Islands Latrunculia Sponge. Org. Lett. 2013, 15, 1516–1519. [Google Scholar] [CrossRef] [PubMed]
- Marialuisa, M.; Anna, A.; Filomena, D.; Concetta, I.; Paolo, L.; Rocco, V.; Carlo, I.; Rita, S. Conithiaquinones A and B, tetracyclic cytotoxic meroterpenes from the mediterranean Ascidian Aplidium conicum. Eur. J. Org. Chem. 2013, 3241–3246. [Google Scholar] [CrossRef]
- Haidy, N.K.; Ding, Y.Q.; Li, X.C.; Daneel, F.; Frank, R.F.; Marc, S. Beyond Polymaxenolide: Cembrane-Africanane terpenoids from the Hybrid Soft Coral Sinularia maxima × S. polydactyla. J. Nat. Prod. 2009, 72, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Wilanfranco, C.T.; Miho, H.; Saki, K.; Shogo, N.; Kazuaki, T.; Tatsuo, N.; Masaru, H. Stereochemical investigations of isochromenones and isobenzofuranones isolated from Leptosphaeria sp. KTC 727. J. Nat. Prod. 2011, 74, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.Q.; Yang, J.W.; Tang, X.C. Huperzine A protects rat pheochromocytoma cells against hydrogen peroxide-induced injury. Neurosci. Lett. 1999, 275, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Tang, X.C. Huperzine B, a novel acetylcholinesterase inhibitor, attenuates hydrogen peroxide induced injury in PC12 cells. Neurosci. Lett. 2000, 292, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Li, S.P.; Zhao, K.J.; Ji, Z.N.; Song, Z.H.; Dong, T.T.X.; Lo, C.K.; Cheung, J.K.H.; Zhu, S.Q.; Tsim, K.W.K. A polysaccharide isolated from Cordyceps sinensis, a traditional Chinese medicine, protects PC12 cells against hydrogen peroxide-induced injury. Life Sci. 2003, 73, 2503–2513. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of salvianolic acid B and salvianolic acid Y are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, J.; Ju, A.; Zhou, D.; Li, D.; Zhou, W.; Geng, W.; Li, B.; Li, L.; Liu, Y.; He, Y.; et al. Salvianolic Acid Y: A New Protector of PC12 Cells against Hydrogen Peroxide-Induced Injury from Salvia officinalis. Molecules 2015, 20, 683-692. https://doi.org/10.3390/molecules20010683
Gong J, Ju A, Zhou D, Li D, Zhou W, Geng W, Li B, Li L, Liu Y, He Y, et al. Salvianolic Acid Y: A New Protector of PC12 Cells against Hydrogen Peroxide-Induced Injury from Salvia officinalis. Molecules. 2015; 20(1):683-692. https://doi.org/10.3390/molecules20010683
Chicago/Turabian StyleGong, Jun, Aichun Ju, Dazheng Zhou, Dekun Li, Wei Zhou, Wanli Geng, Bing Li, Li Li, Yanjie Liu, Ying He, and et al. 2015. "Salvianolic Acid Y: A New Protector of PC12 Cells against Hydrogen Peroxide-Induced Injury from Salvia officinalis" Molecules 20, no. 1: 683-692. https://doi.org/10.3390/molecules20010683





