Crystallographic Studies Evidencing the High Energy Tolerance to Disrupting the Interface Disulfide Bond of Thioredoxin 1 from White Leg Shrimp Litopenaeus vannamei
Abstract
:1. Introduction
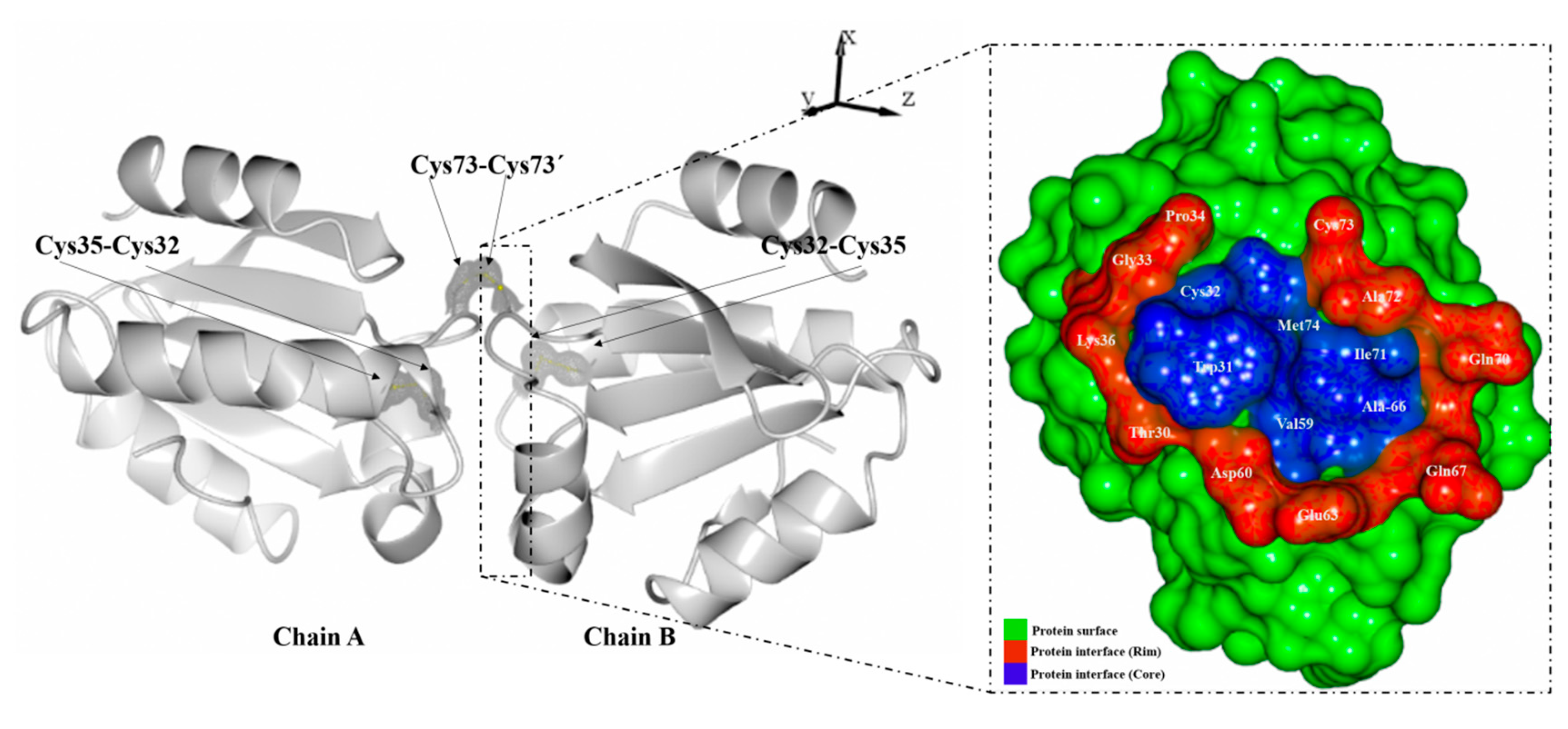
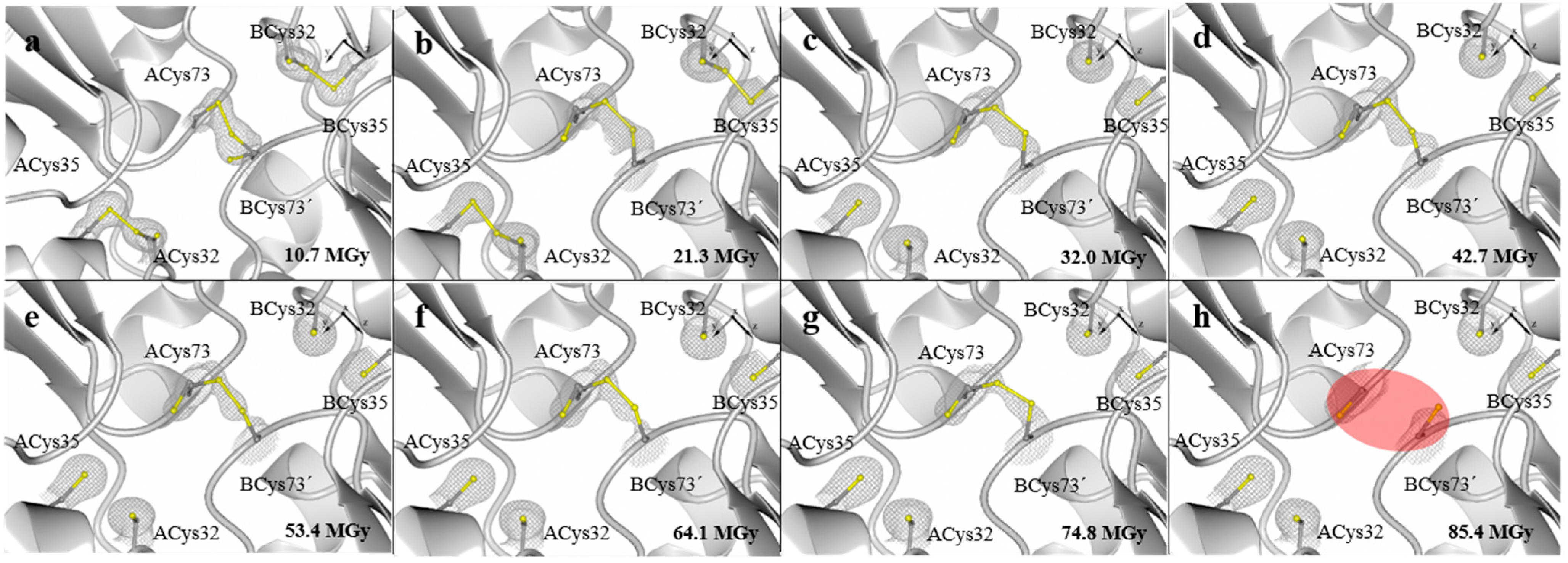
2. Results and Discussion

3. Experimental Section
3.1. Experimental Procedures
3.2. Data Collection and X-ray Diffraction Experiments
3.3. Data Processing, Molecular Replacement and Refinement
| Parameters | LvTrx-1x (1) | LvTrx-1x (2) | LvTrx-1x (3) | LvTrx-1x (6) | LvTrx-1x (7) | LvTrx-1x (8) | LvTrx-1x (9) | LvTrx-1x (10) |
|---|---|---|---|---|---|---|---|---|
| Data Collection Statistics | ||||||||
| X-ray source | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 |
| Wavelength (Å) | 0.9686 | 0.9686 | 0.9686 | 0.9686 | 0.9686 | 0.9686 | 0.9686 | 0.9686 |
| Space group | P3212 | P3212 | P3212 | P3212 | P3212 | P3212 | P3212 | P3212 |
| Absorbed dose (MGy) | 3.4 | 6.8 | 10.3 | 20.6 | 24.0 | 27.4 | 30.9 | 34.3 |
| Unit-cell dimensions | ||||||||
| a, b, c (Å) | 57.18, 57.18, 117.62 | 57.21, 57.21, 117.75 | 57.24, 57.24, 117.85 | 57.44, 57.44, 117.90 | 57.41, 57.41, 117.85 | 57.36, 57.36, 117.85 | 57.46, 57.46, 117.90 | 57.21, 57.21, 117.95 |
| α, β, γ angles (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution range (Å) | 28.59–1.65 | 28.60–1.71 | 28.61–1.65 | 45.83–1.72 | 45.80–1.74 | 45.75–1.76 | 45.85–1.80 | 27.80–1.84 |
| No. of reflections | 86,984 | 76,685 | 84,052 | 63,098 | 61,524 | 61,048 | 61,404 | 61,761 |
| No. of unique reflections | 26,586 | 23,647 | 25,952 | 23,101 | 22,342 | 21,661 | 20,239 | 18,958 |
| Completeness (%) | 99.9 (99.8) | 98.0 (91.6) | 98.2 (92.8) | 96.7 (95.5) | 97.0 (96.1) | 97.3 (95.7) | 96.6 (96.4) | 97.3 (90.7) |
| Rsvm (%) § | 3.8 (43.9) | 3.4 (34.3) | 3.7 (50.5) | 6.9 (39.1) | 6.9 (39.6) | 7.6 (40) | 5.2 (40.4) | 4.8 (42.4) |
| I/σ(I) | 15.9 (2.4) | 18.0 (2.9) | 15.0 (2.0) | 7.5 (2.0) | 7.7 (2.0) | 7.3 (2.0) | 7.2 (1.36) | 13.0 (2.8) |
| Multiplicity | 3.3 (3.3) | 3.2 (3.0) | 3.2 (3.0) | 2.7 (2.5) | 2.8 (2.6) | 2.8 (2.6) | 2.5 (2.2) | 3.3 (3.2) |
| Asymmetric unit | Dimer | Dimer | Dimer | Dimer | Dimer | Dimer | Dimer | Dimer |
| Refinement Statistics | ||||||||
| Rwork/Rfree (%) | 18.87/22.33 | 18.39/23.41 | 19.22/23.15 | 18.77/22.98 | 18.76/23.72 | 18.66/23.93 | 18.66/23.37 | 18.45/21.72 |
| B-value (Å2) | ||||||||
| Protein | 25.2 | 26.8 | 30.4 | 25.53 | 26.31 | 25.05 | 25.64 | 28.52 |
| Ion/Ligand | 32.21 | 35.63 | 38.31 | 32.69 | 34.35 | 35.97 | 35.47 | 51.61 |
| Water | 37.69 | 38.98 | 41.64 | 37.68 | 37.46 | 36.74 | 36.75 | 39.21 |
| All atoms | 31.63 | 33.80 | 36.78 | 31.96 | 32.70 | 32.58 | 32.62 | 39.78 |
| Wilson plot B-value (Å2) | 24.97 | 25.8 | 26.46 | 25.93 | 26.67 | 27.21 | 27.90 | 30.01 |
| RMSD from Ideal Stereochemistry | ||||||||
| Bond lengths (Å) | 0.005 | 0.005 | 0.005 | 0.005 | 0.005 | 0.004 | 0.005 | 0.005 |
| Bond angles (°) | 0.924 | 0.899 | 0.921 | 0.940 | 0.932 | 0.816 | 0.867 | 0.885 |
| Coordinate error (maximum-likelihood base) (Å) | 0.19 | 0.23 | 0.20 | 0.26 | 0.25 | 0.26 | 0.24 | 0.21 |
| Ramachandran plot (%) | ||||||||
| Most-favored regions | 97.75 | 97.78 | 97.32 | 97.32 | 97.77 | 97.32 | 97.31 | 97.75 |
| Additional allowed regions | 2.25 | 2.22 | 2.68 | 2.68 | 2.23 | 2.68 | 2.69 | 2.25 |
| Disallowed regions | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Parameters | LvTrx-3x (1) | LvTrx-3x (2) | LvTrx-3x (3) | LvTrx-3x (4) | LvTrx-3x (5) | LvTrx-3x (6) | LvTrx-3x (7) | LvTrx-3x (8) |
|---|---|---|---|---|---|---|---|---|
| Data Collection Statistics | ||||||||
| X-ray source | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 | DLS Beamline I24 |
| Wavelength (Å) | 0.9686 | 0.9686 | 0.9686 | 0.9686 | 0.9686 | 0.9686 | 0.9686 | 0.9686 |
| Space group | P3212 | P3212 | P3212 | P3212 | P3212 | P3212 | P3212 | P3212 |
| Absorbed dose (MGy) | 10.7 | 21.3 | 32.0 | 42.7 | 53.4 | 64.1 | 74.8 | 85.4 |
| Unit-cell dimensions | ||||||||
| a, b, c (Å) | 57.49, 57.49, 117.92 | 57.55, 57.55, 118.02 | 57.61, 57.61, 118.05 | 57.65, 57.65, 118.06 | 57.68, 57.68, 118.05 | 57.70, 57.70, 118.05 | 57.69, 57.69, 118.02 | 57.70, 57.70, 118.02 |
| α, β, γ angles (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution range (Å) | 28.75–1.46 | 27.95–1.60 | 28.80–1.70 | 25.90–1.80 | 28.01–1.88 | 28.02–1.96 | 28.02–2.05 | 28.85–2.15 |
| No. of reflections | 108,975 | 97,790 | 81,543 | 69,057 | 60,539 | 53,701 | 46,718 | 40,484 |
| No. of unique reflections | 38,937 | 29,741 | 24,912 | 21,055 | 18,546 | 16,387 | 14,356 | 12,470 |
| Completeness (%) | 99.6 (99.4) | 99.6 (99.4) | 99.6 (99.6) | 99.8 (99.6) | 99.8 (99.6) | 99.9 (99.6) | 99.6 (99.7) | 99.6 (99.6) |
| Rsym(%) § | 2.5 (38.4) | 2.7 (39.9) | 2.9 (44.0) | 3.3 (40.5) | 3.9 (42.1) | 4.5 (45) | 4.8 (42.6) | 5.0 (36.7) |
| I/σ(I) | 21.8 (3.2) | 21.5 (3.1) | 21.1 (2.8) | 19.2 (3.1) | 16.7 (2.9) | 14.9 (2.9) | 14.4 (3.0) | 14.1 (3.5) |
| Multiplicity | 3.3 (3.2) | 3.3 (3.3) | 3.3 (3.2) | 3.3 (3.3) | 3.3 (3.2) | 3.3 (3.2) | 3.3 (3.3) | 3.2 (3.2) |
| Asymmetric unit | Dimer | Dimer | Dimer | Dimer | Dimer | Dimer | Dimer | Dimer |
| Refinement Statistics | ||||||||
| Rwork/Rfree(%) | 19.95/22.02 | 18.67/23.36 | 18.71/20.67 | 18.58/22.65 | 18.23/22.63 | 17.78/21.72 | 18.66/23.37 | 18.03/24.63 |
| B-value (Å2) | ||||||||
| Protein | 24.83 | 23.88 | 28.13 | 29.22 | 31.2 | 34.54 | 34.29 | 36.59 |
| Ion/Ligand | 47.50 | 42.28 | 52.88 | 51.69 | 55.57 | 61.46 | 52.27 | 64.22 |
| Water | 38.0 | 36.65 | 40.39 | 41.02 | 41.61 | 43.77 | 42.05 | 44.55 |
| All atoms | 36.77 | 34.27 | 40.46 | 40.64 | 42.79 | 46.65 | 42.87 | 48.45 |
| Wilson plot B-value (Å2) | 20.55 | 23.61 | 26.69 | 29.43 | 31.99 | 34.28 | 36.30 | 37.78 |
| RMSD from Ideal Stereochemistry | ||||||||
| Bond lengths (Å) | 0.004 | 0.006 | 0.004 | 0.004 | 0.005 | 0.004 | 0.004 | 0.006 |
| Bond angles (°) | 0.861 | 1.027 | 0.824 | 0.826 | 0.932 | 0.842 | 0.756 | 0.938 |
| Coordinate error (maximum-likelihood base) (Å) | 0.18 | 0.18 | 0.17 | 0.21 | 0.23 | 0.19 | 0.24 | 0.25 |
| Ramachandran plot (%) | ||||||||
| Most-favored regions | 97.77 | 97.32 | 97.75 | 97.30 | 98.20 | 97.75 | 98.19 | 97.75 |
| Additional allowed regions | 2.23 | 2.68 | 2.25 | 2.70 | 1.80 | 2.25 | 1.81 | 2.25 |
| Disallowed regions | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Murray, J.W.; Rudiño-Piñera, E.; Owen, R.L.; Grininger, M.; Ravelli, R.B.G.; Garman, E.F. Parameters affecting the X-ray dose absorbed by macromolecular crystals. J. Synchrotron Radiat. 2005, 12, 268–275. [Google Scholar] [PubMed]
- Weik, M.; Ravelli, R.B.; Kryger, G.; McSweeney, S.; Raves, M.L.; Harel, M.; Gros, P.; Silman, I.; Kroon, J.; Sussman, J.L. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc. Natl. Acad. Sci. USA 2000, 97, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Adam, V.; Royant, A.; Nivière, V.; Molina-Heredia, F.P.; Bourgeois, D. Structure of superoxide reductase bound to ferrocyanide and active site expansion upon X-ray-induced photo-reduction. Structure 2004, 12, 1729–1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burmeister, W.P. Structural changes in a cryo-cooled protein crystal owing to radiation damage. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000, 56, 328–341. [Google Scholar] [CrossRef]
- Ravelli, R.B.; McSweeney, S.M. The “fingerprint” that X-rays can leave on structures. Structure 2000, 8, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Fioravanti, E.; Vellieux, F.M.D.; Amara, P.; Madern, D.; Weik, M. Specific radiation damage to acidic residues and its relation to their chemical and structural environment. J. Synchrotron Radiat. 2007, 14, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Petrova, T.; Ginell, S.; Mitschler, A.; Kim, Y.; Lunin, V.Y.; Joachimiak, G.; Cousido-Siah, A.; Hazemann, I.; Podjarny, A.; Lazarski, K.; et al. X-ray-induced deterioration of disulfide bridges at atomic resolution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 1075–1091. [Google Scholar]
- Henderson, R. Cryo-Protection of Protein Crystals against Radiation Damage in Electron and X-ray Diffraction. Proc. R. Soc. B Biol. Sci. 1990, 241, 6–8. [Google Scholar] [CrossRef]
- Owen, R.L.; Rudiño-Piñera, E.; Garman, E.F. Experimental determination of the radiation dose limit for cryocooled protein crystals. Proc. Natl. Acad. Sci. USA 2006, 103, 4912–4917. [Google Scholar] [CrossRef] [PubMed]
- Garman, E.F.; Owen, R.L. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Chinte, U.; Shah, B.; Chen, Y.-S.; Pinkerton, A.A.; Schall, C.A.; Hanson, B.L. Cryogenic (<20 K) helium cooling mitigates radiation damage to protein crystals. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 63, 486–492. [Google Scholar]
- Petrova, T.; Lunin, V.Y.; Ginell, S.; Hazemann, I.; Lazarski, K.; Mitschler, A.; Podjarny, A.; Joachimiak, A. X-ray-radiation-induced cooperative atomic movements in protein. J. Mol. Biol. 2009, 387, 1092–1105. [Google Scholar] [CrossRef] [PubMed]
- Teng, T.Y.; Moffat, K. Radiation damage of protein crystals at cryogenic temperatures between 40 K and 150 K. J. Synchrotron Radiat. 2002, 9, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Campos-Acevedo, A.A.; Garcia-Orozco, K.D.; Sotelo-Mundo, R.R.; Rudiño-Piñera, E. Expression, purification, crystallization and X-ray crystallographic studies of different redox states of the active site of thioredoxin 1 from the whiteleg shrimp Litopenaeus vannamei. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Saha, R.P.; Bahadur, R.P.; Pal, A.; Mandal, S.; Chakrabarti, P. ProFace: a server for the analysis of the physicochemical features of protein-protein interfaces. BMC Struct. Biol. 2006, 6. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Weichsel, A.; Kem, M.; Montfort, W.R. Crystal structure of human thioredoxin revealing an unraveled helix and exposed S-nitrosation site. Protein Sci. 2010, 19, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Gronenborn, A.M.; Clore, G.M.; Louis, J.M.; Wingfield, P.T. Is human thioredoxin monomeric or dimeric? Protein Sci. 1999, 8, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Orozco, K.D.; Sanchez-Paz, A.; Aispuro-Hernandez, E.; Gomez-Jimenez, S.; Lopez-Zavala, A.; Araujo-Bernal, S.; Muhlia-Almazan, A. Gene expression and protein levels of thioredoxin in the gills from the whiteleg shrimp (Litopenaeus vannamei) infected with two different viruses: The WSSV or IHHNV. Fish Shellfish Immunol. 2012, 32, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.W.; Garman, E.F.; Ravelli, R.B.G. X-ray absorption by macromolecular crystals: The effects of wavelength and crystal composition on absorbed dose. J. Appl. Crystallogr. 2004, 37, 513–522. [Google Scholar] [CrossRef]
- Paithankar, K.S.; Owen, R.L.; Garman, E.F. Absorbed dose calculations for macromolecular crystals: Improvements to RADDOSE. J. Synchrotron Radiat. 2009, 16, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Paithankar, K.S.; Garman, E.F. Know your dose: RADDOSE. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Sutton, K.A.; Black, P.J.; Mercer, K.R.; Garman, E.F.; Owen, R.L.; Snell, E.H.; Bernhard, W.A. Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 2381–2394. [Google Scholar] [CrossRef] [PubMed]
- Leiros, H.-K.S.; McSweeney, S.M.; Smalås, A.O. Atomic resolution structures of trypsin provide insight into structural radiation damage. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001, 57, 488–497. [Google Scholar] [CrossRef]
- Holmgren, A. Thioredoxin: Structure and functions. Trends Biochem. Sci. 1981, 6, 26–29. [Google Scholar] [CrossRef]
- Lee, B.; Richards, F.M. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 1971, 55, 379–400. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 72–82. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Katti, S.K.; LeMaster, D.M.; Eklund, H. Crystal structure of thioredoxin from Escherichia coli at 1.68 A resolution. J. Mol. Biol. 1990, 212, 167–184. [Google Scholar] [CrossRef]
- Andersen, J.F.; Sanders, D.A.; Gasdaska, J.R.; Weichsel, A.; Powis, G.; Montfort, W.R. Human thioredoxin homodimers: Regulation by pH, role of aspartate 60, and crystal structure of the aspartate 60 → asparagine mutant. Biochemistry 1997, 36, 13979–13988. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Sample of Thioredoxin 1 from Litopenaeus vannamei is available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos-Acevedo, A.A.; Rudiño-Piñera, E. Crystallographic Studies Evidencing the High Energy Tolerance to Disrupting the Interface Disulfide Bond of Thioredoxin 1 from White Leg Shrimp Litopenaeus vannamei. Molecules 2014, 19, 21113-21126. https://doi.org/10.3390/molecules191221113
Campos-Acevedo AA, Rudiño-Piñera E. Crystallographic Studies Evidencing the High Energy Tolerance to Disrupting the Interface Disulfide Bond of Thioredoxin 1 from White Leg Shrimp Litopenaeus vannamei. Molecules. 2014; 19(12):21113-21126. https://doi.org/10.3390/molecules191221113
Chicago/Turabian StyleCampos-Acevedo, Adam A., and Enrique Rudiño-Piñera. 2014. "Crystallographic Studies Evidencing the High Energy Tolerance to Disrupting the Interface Disulfide Bond of Thioredoxin 1 from White Leg Shrimp Litopenaeus vannamei" Molecules 19, no. 12: 21113-21126. https://doi.org/10.3390/molecules191221113




