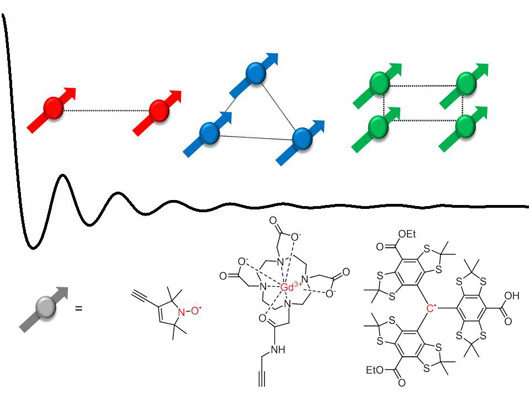
Strategies for the Synthesis of Yardsticks and Abaci for Nanometre Distance Measurements by Pulsed EPR
Abstract
:1. Introduction
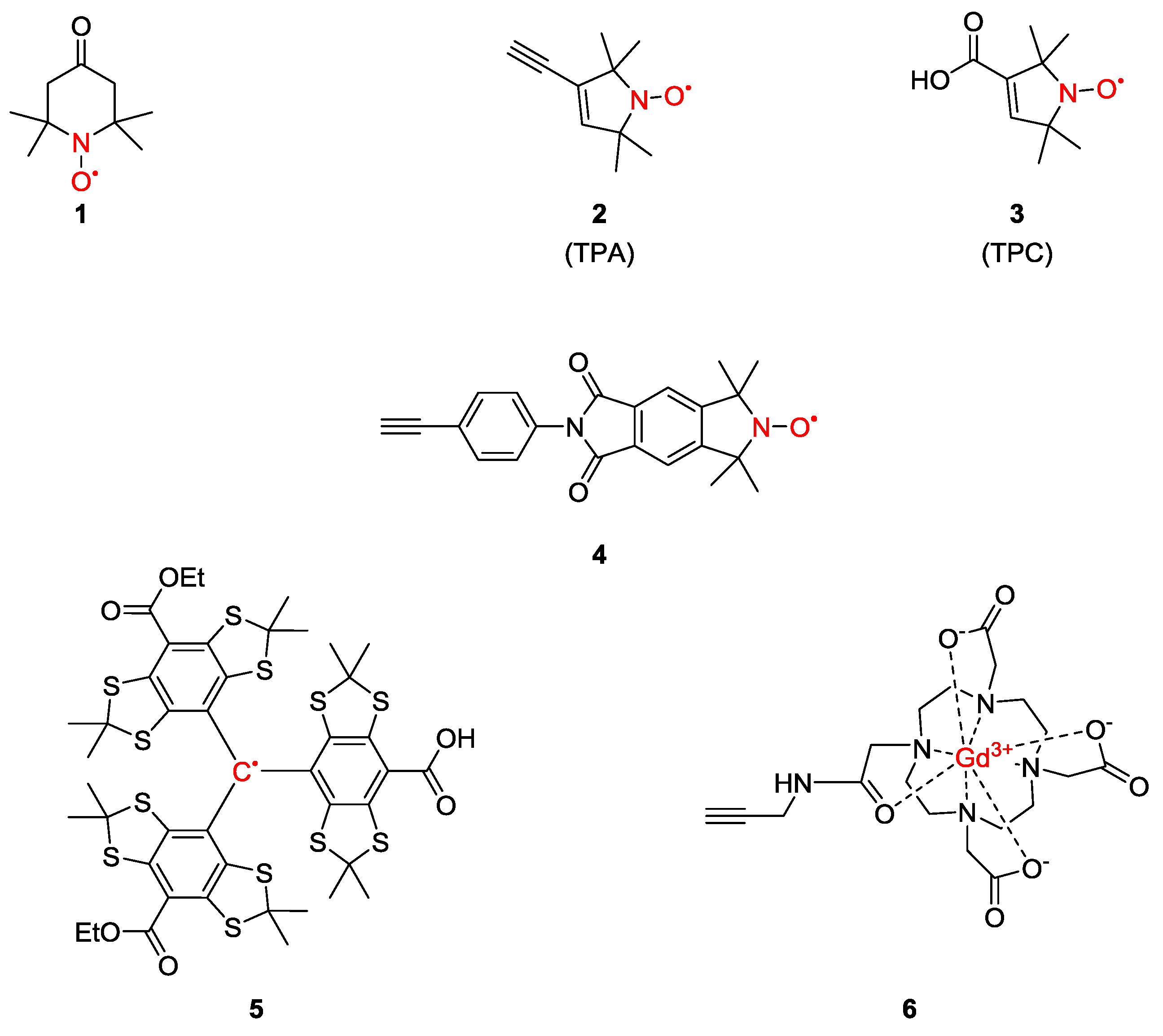
2. Spin-Labels
3. Model Systems
3.1. Biradicals
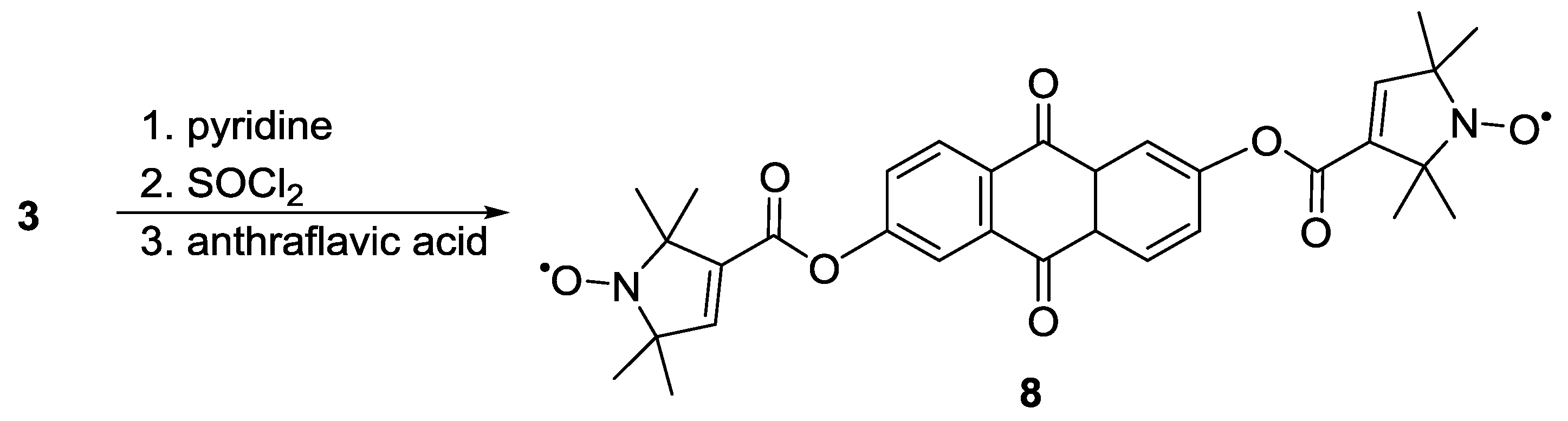
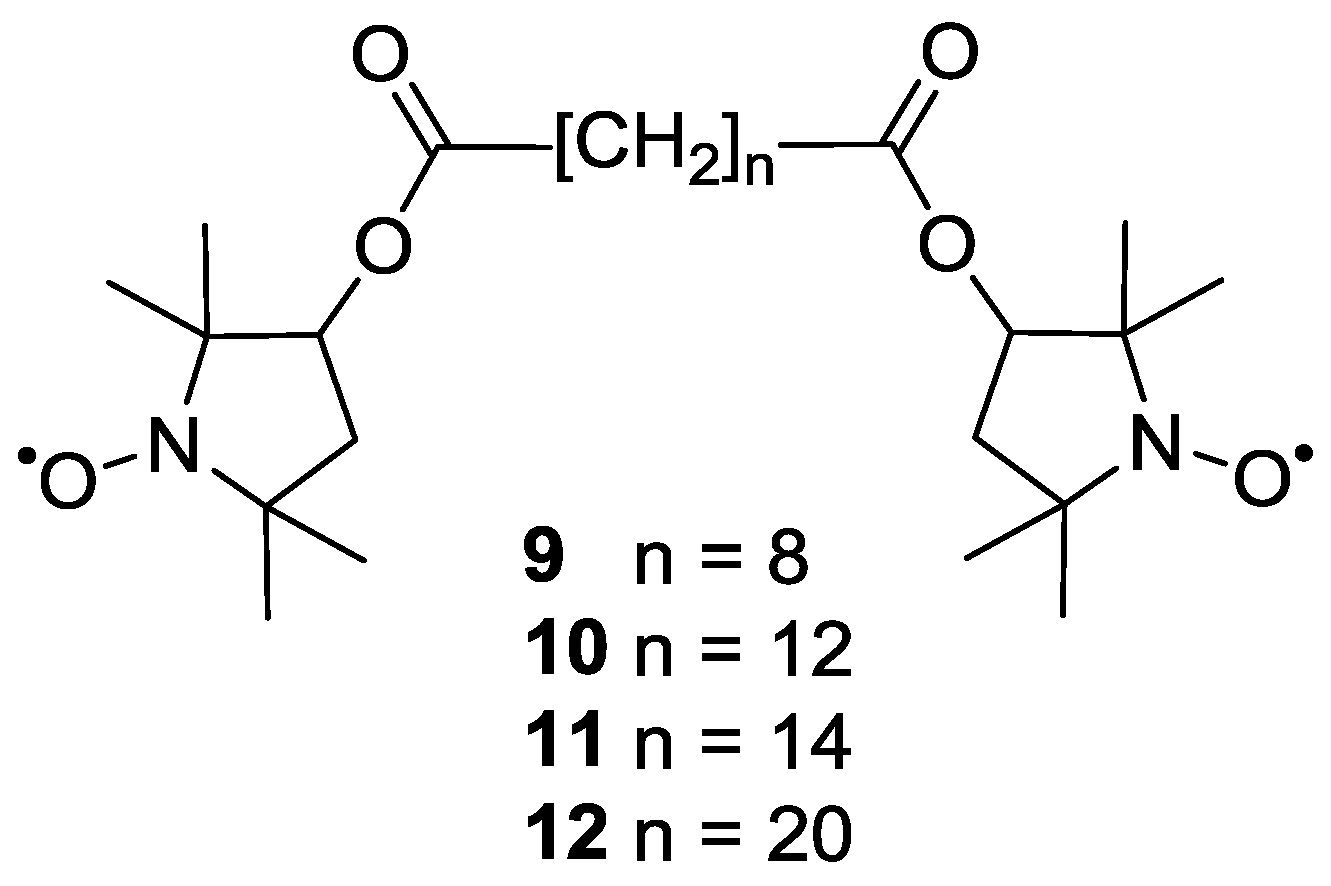
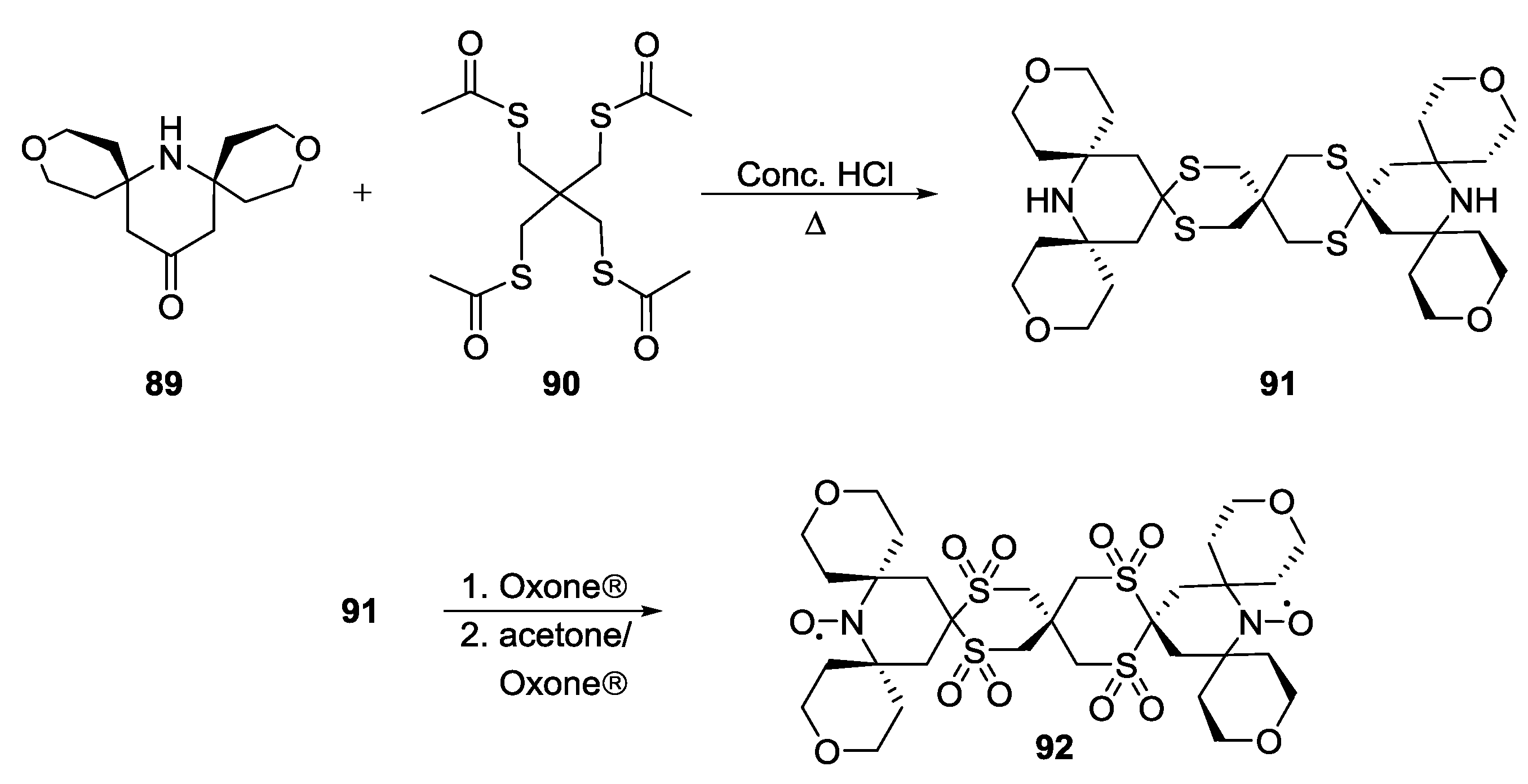
3.1.1. Nitroxide-Based Biradicals

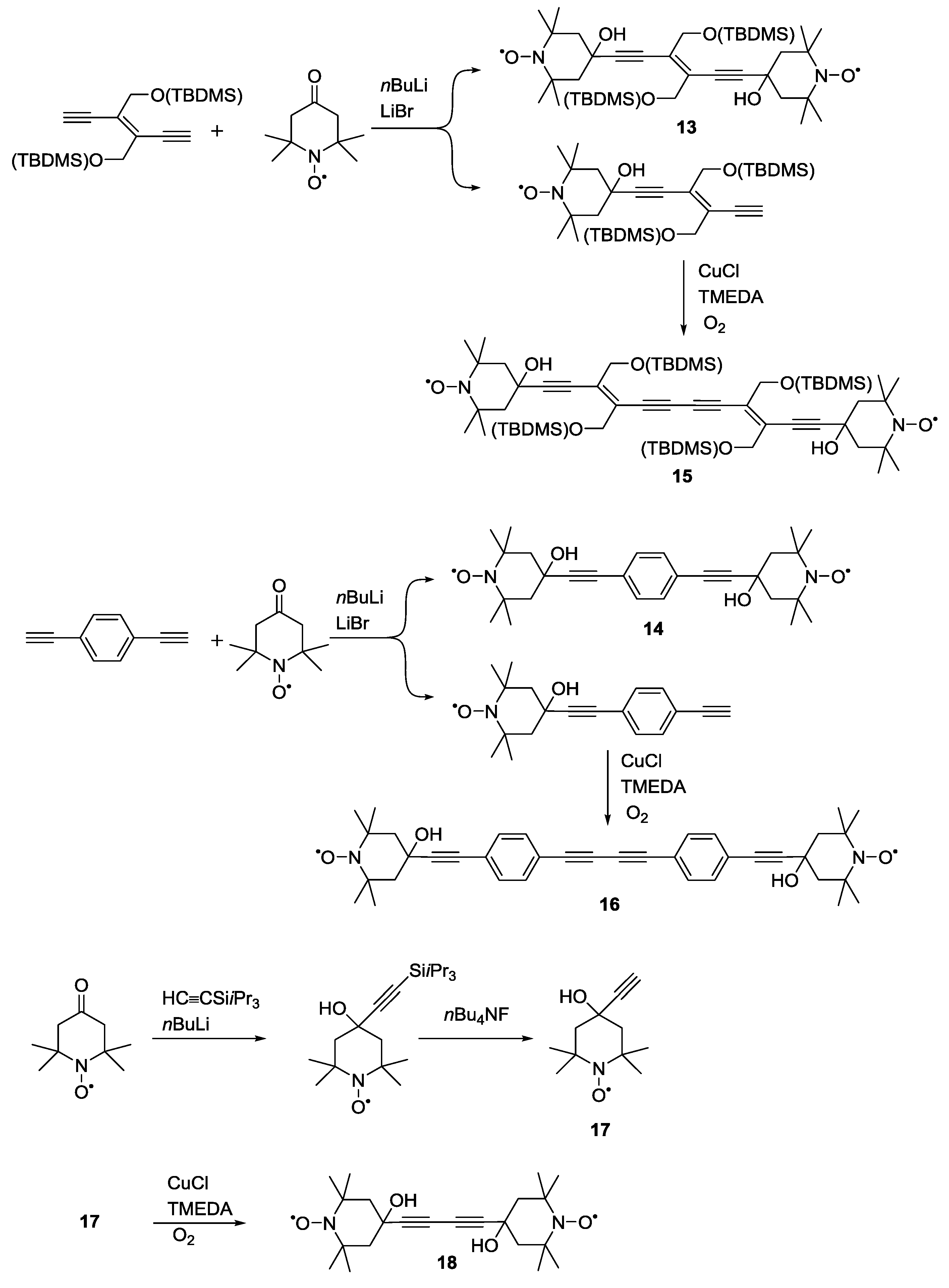
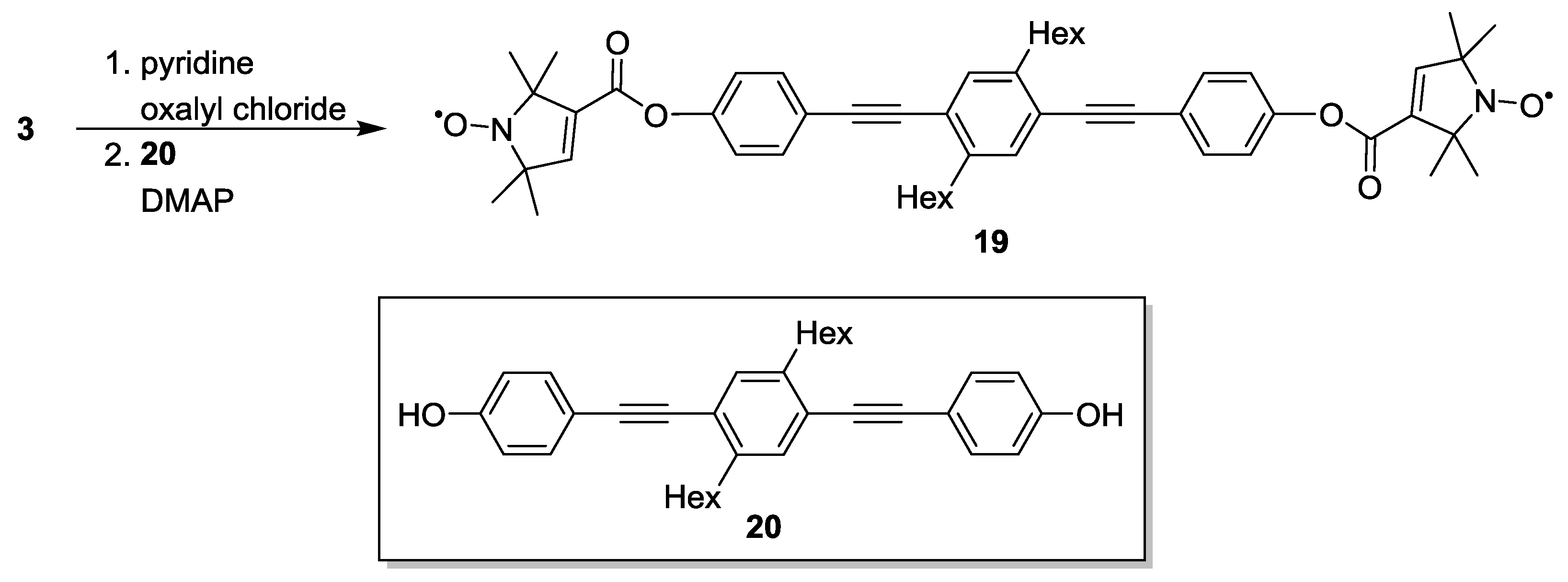
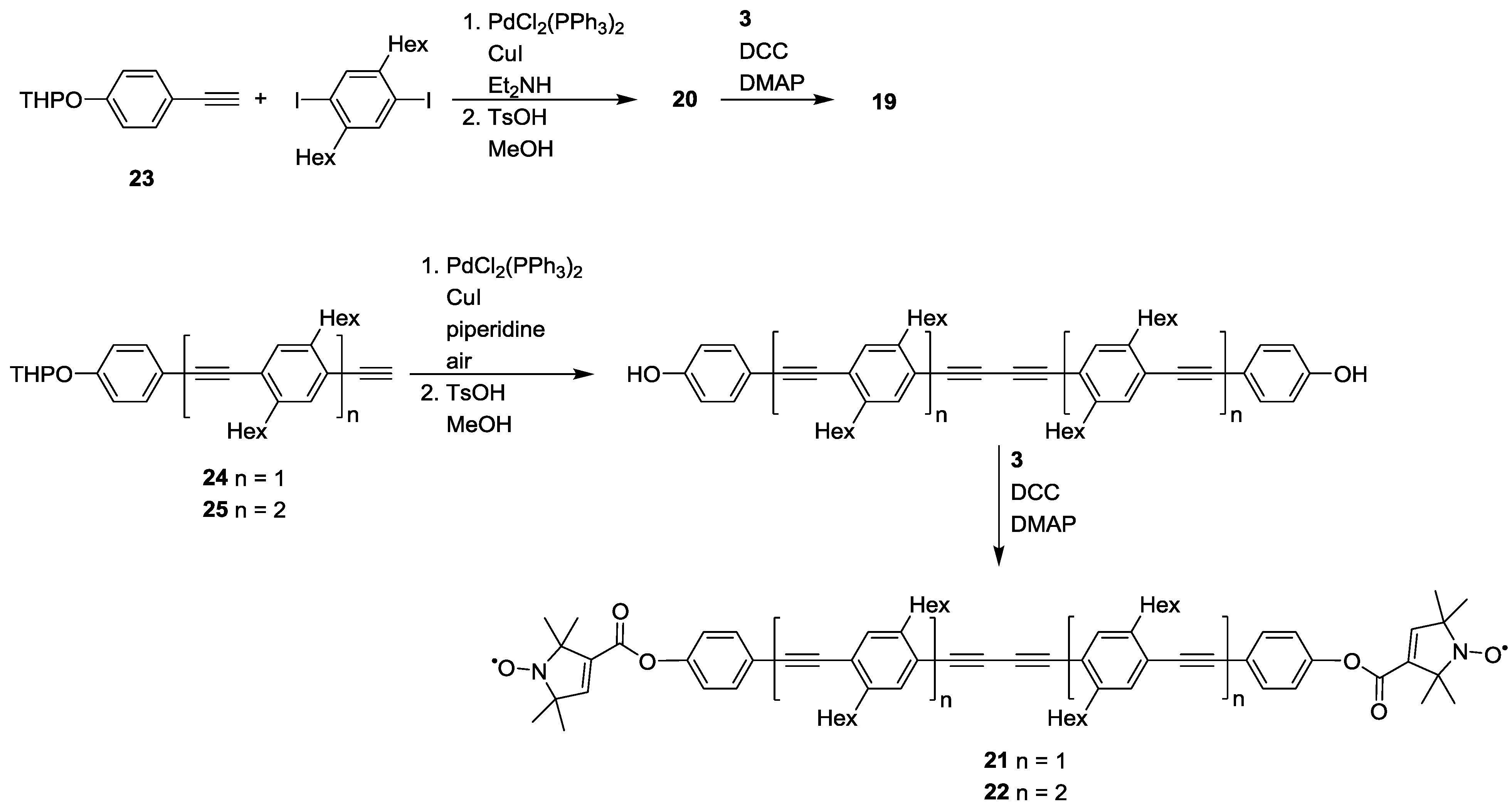
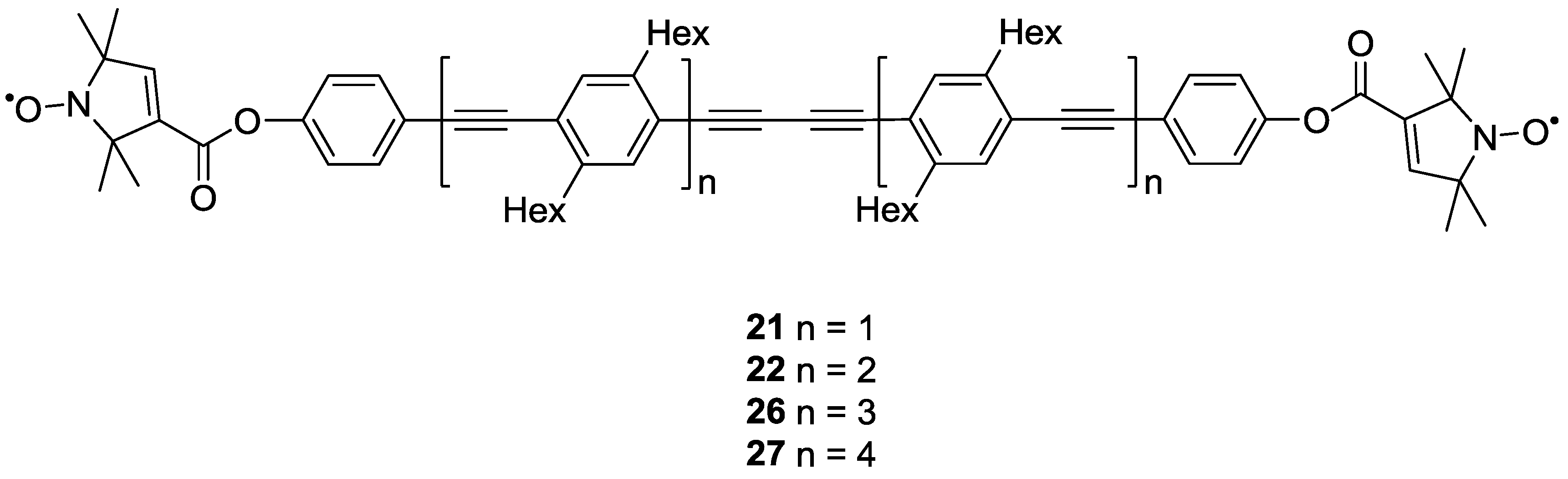
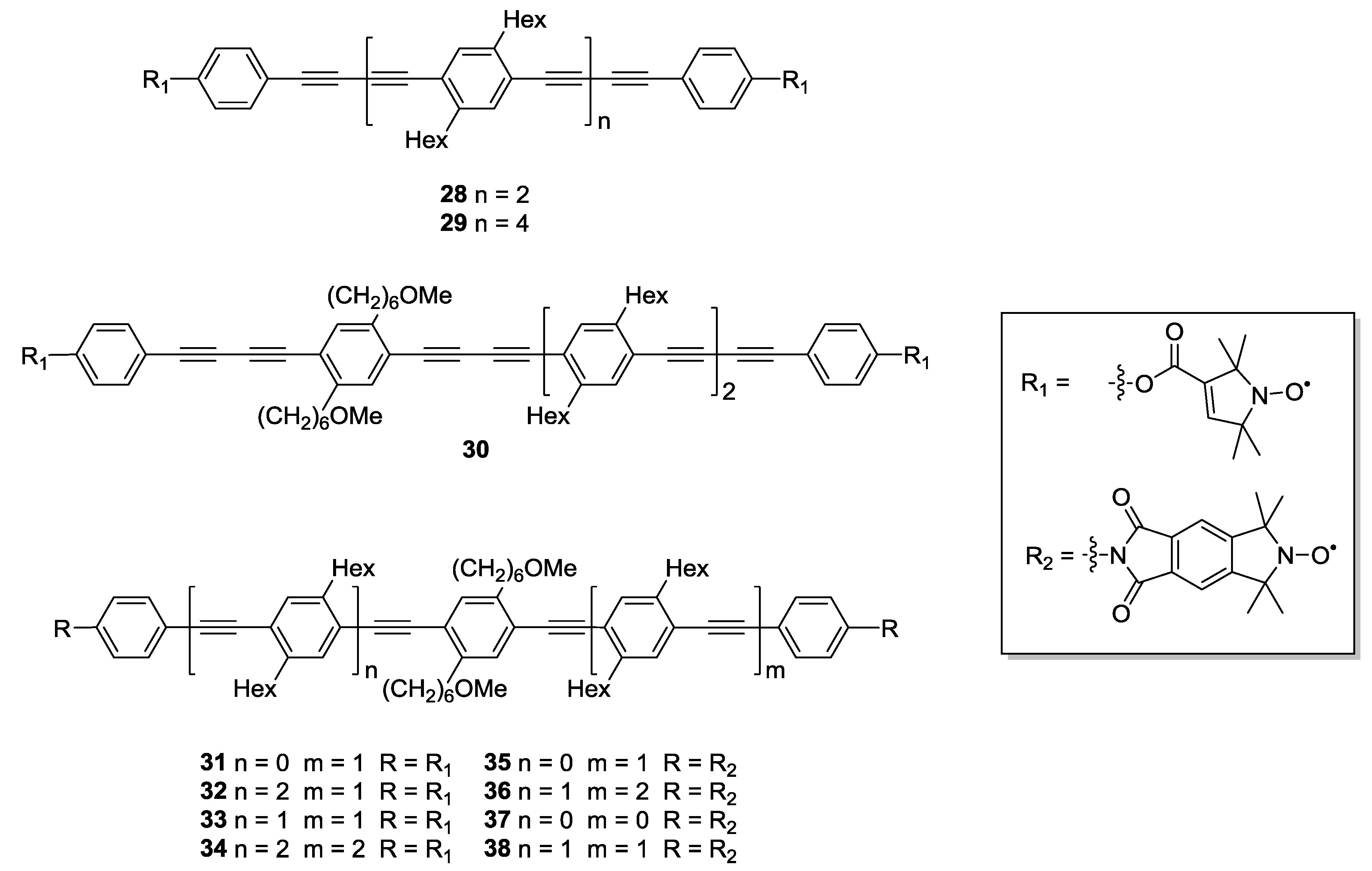
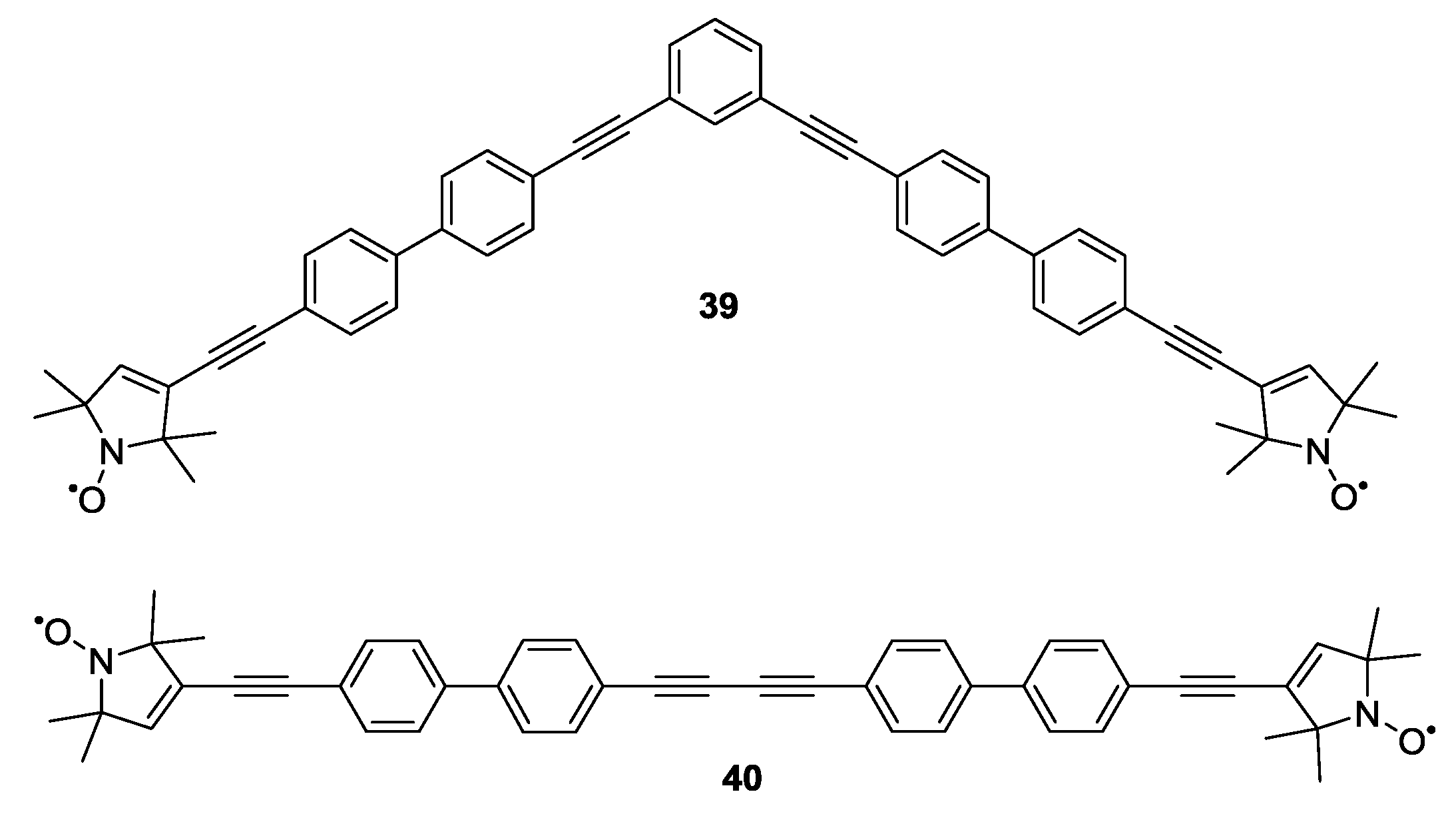
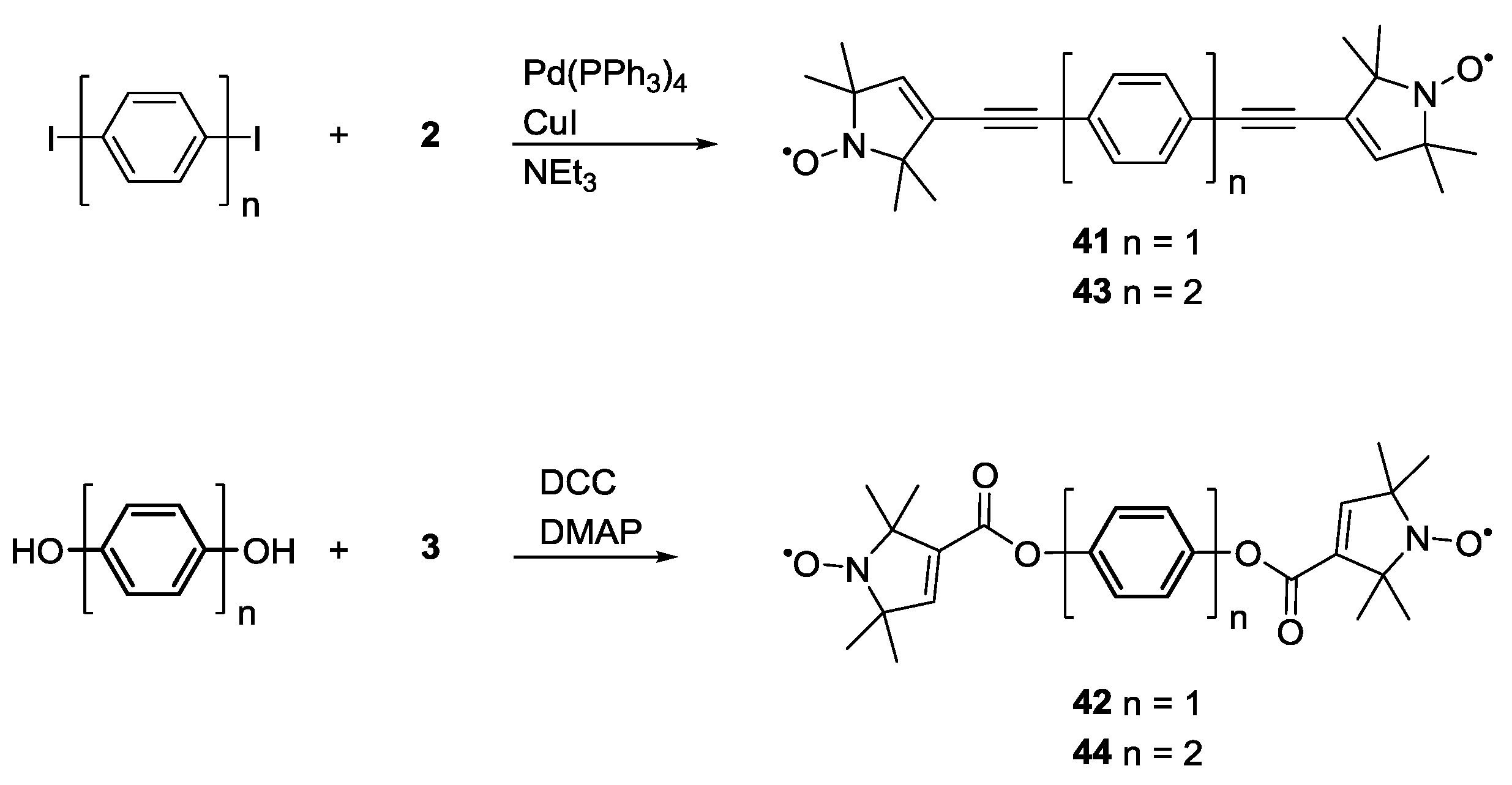
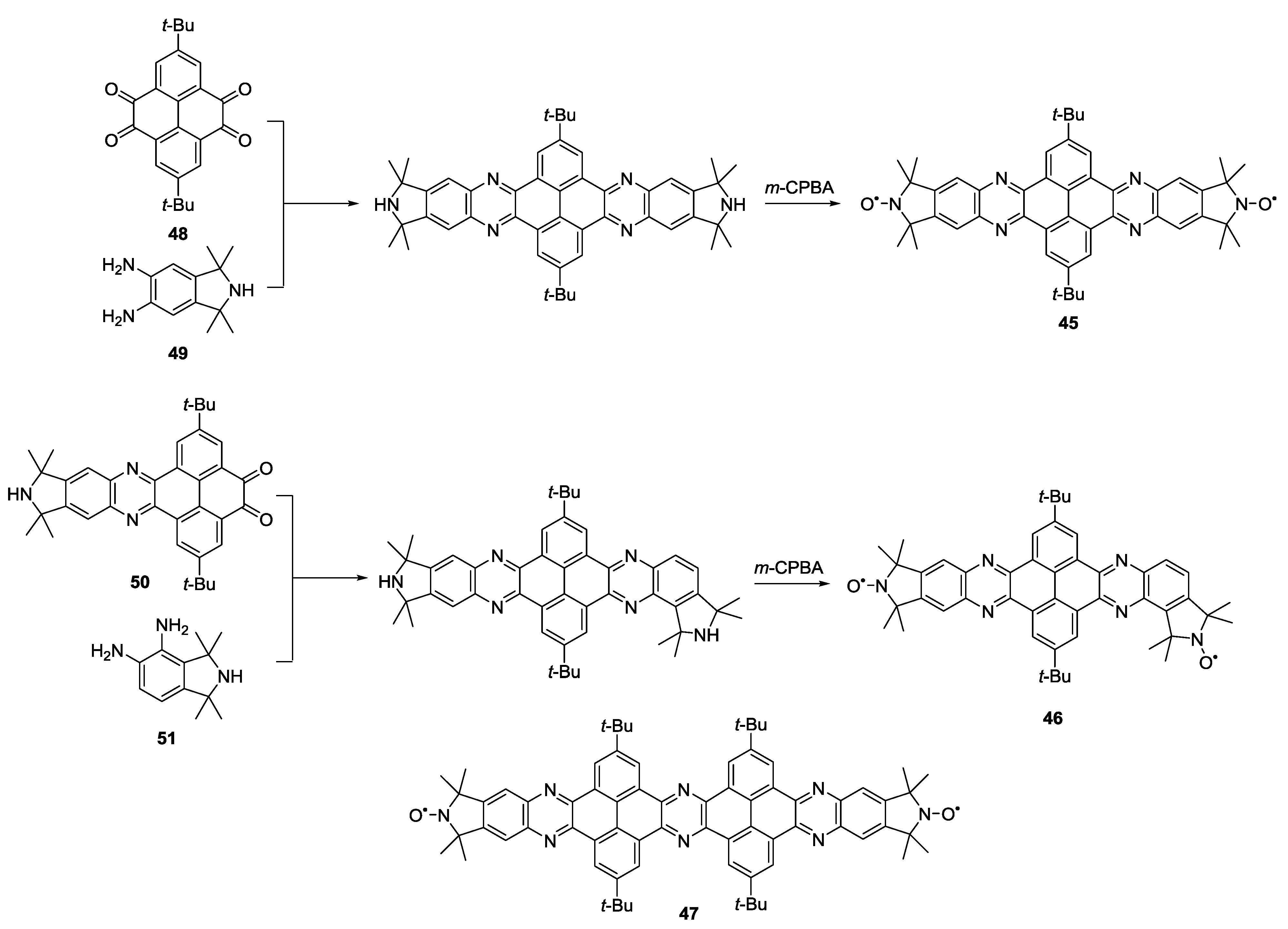
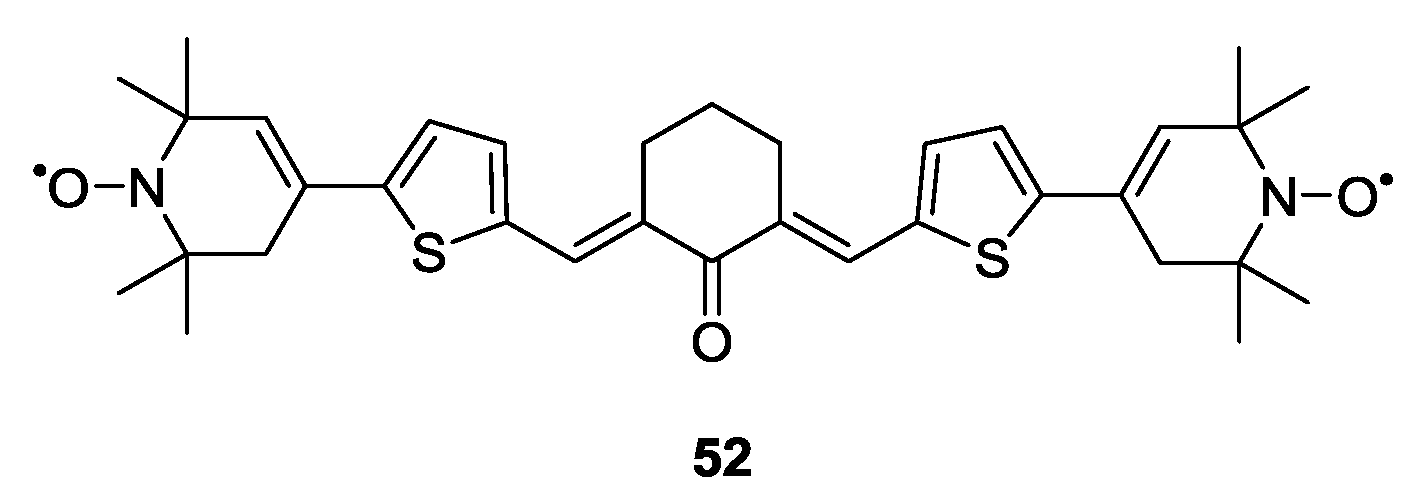
3.1.2. Trityl-Based Biradicals
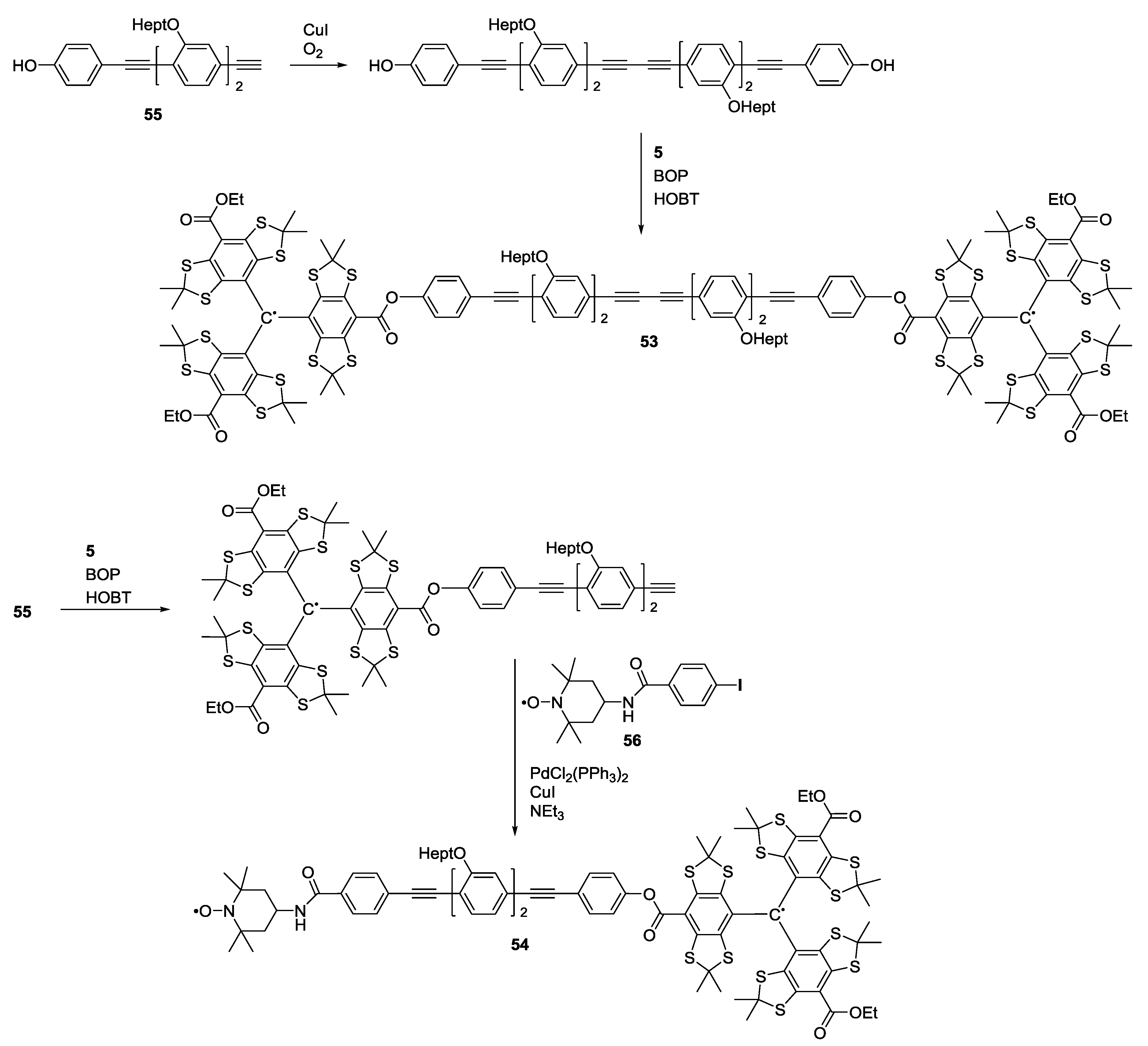
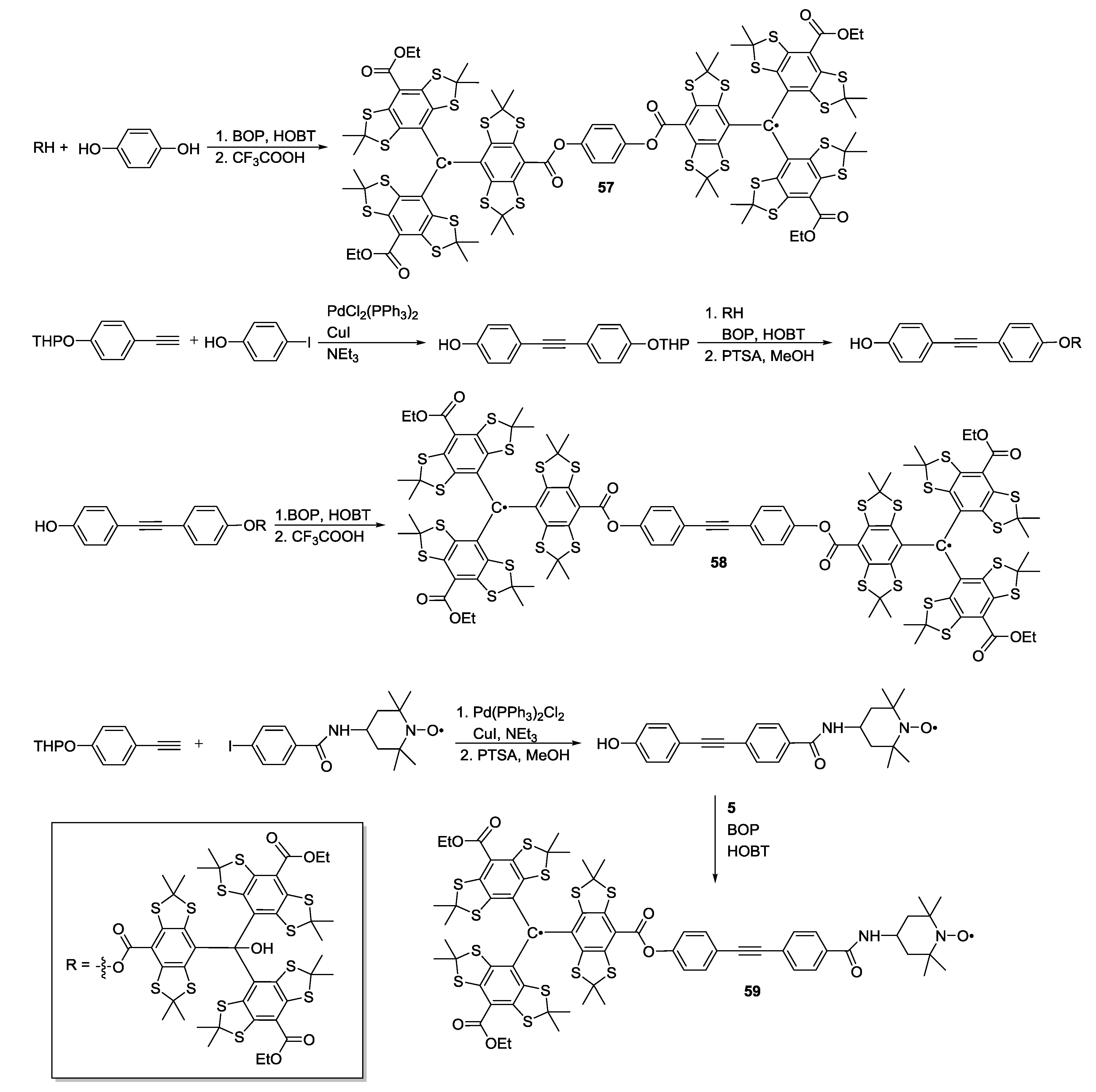
3.2. Polyradicals
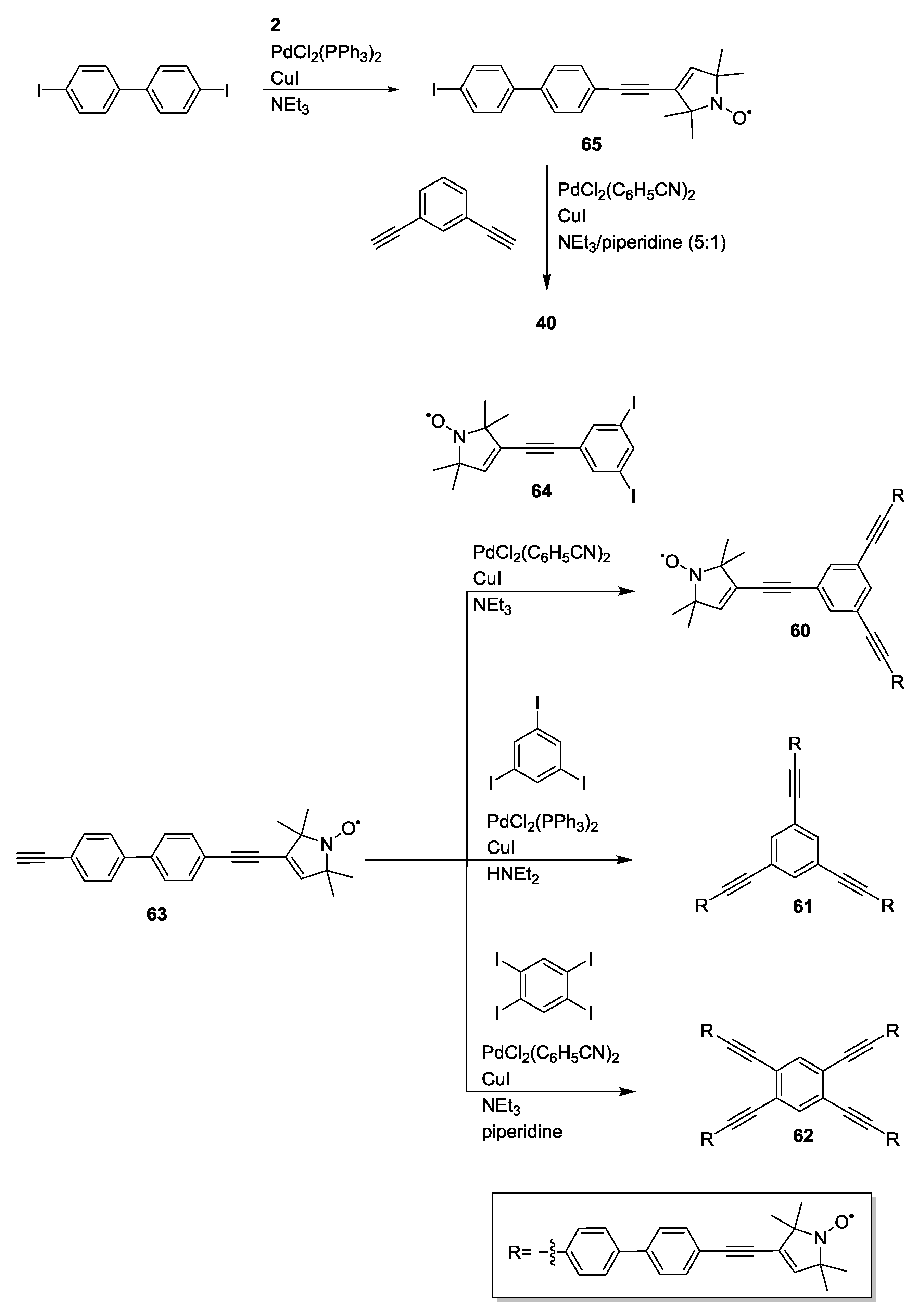
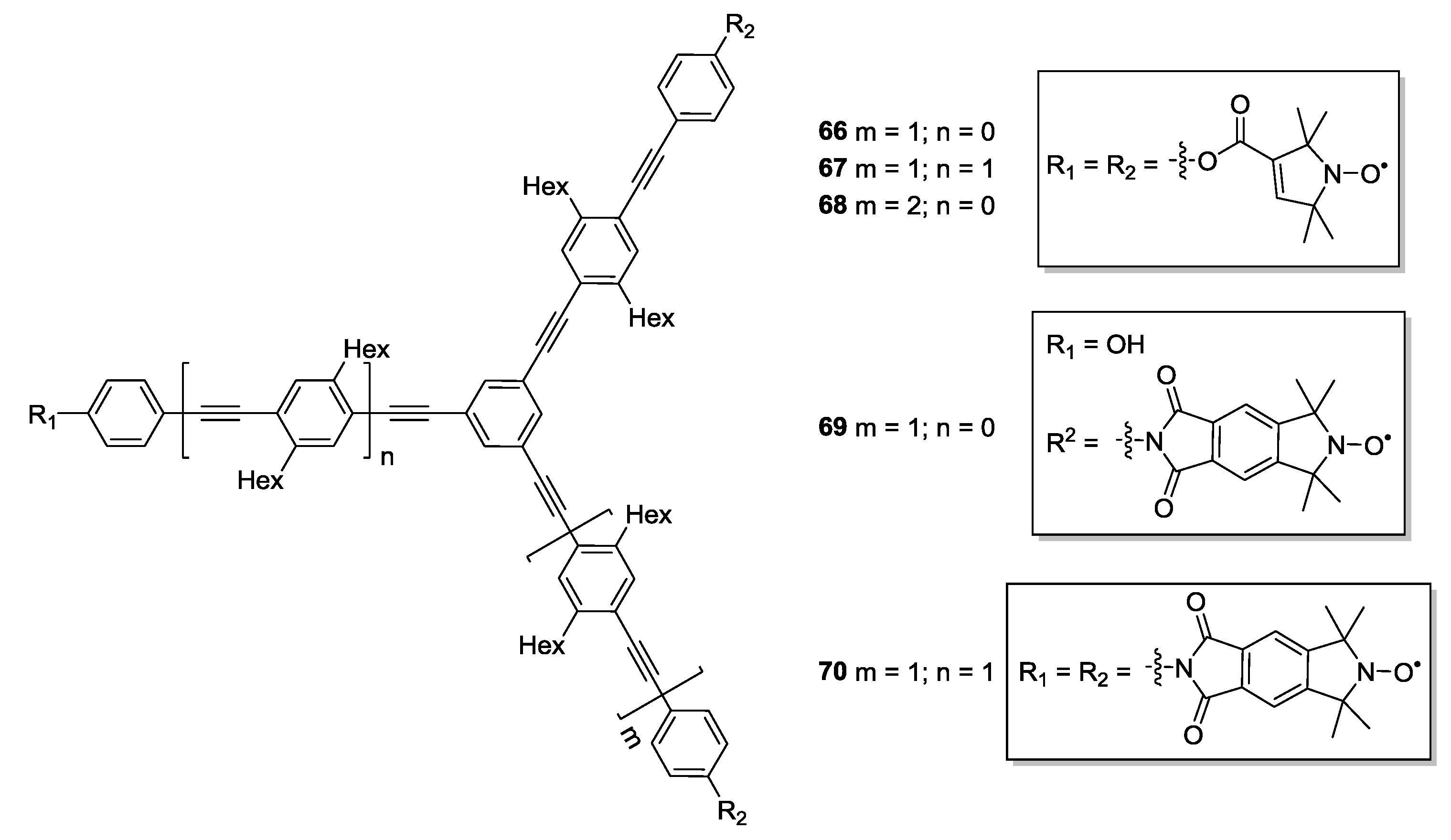
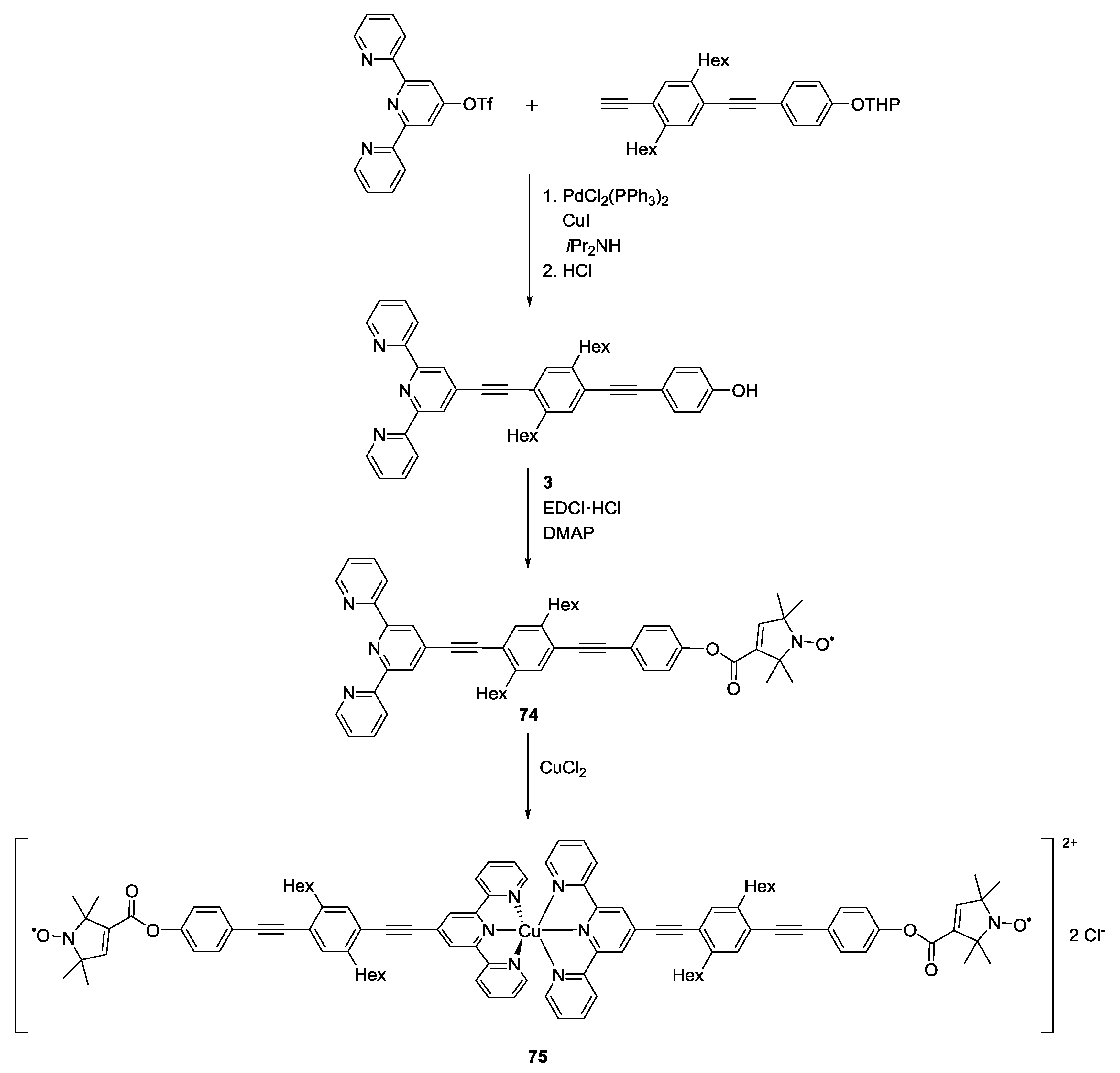
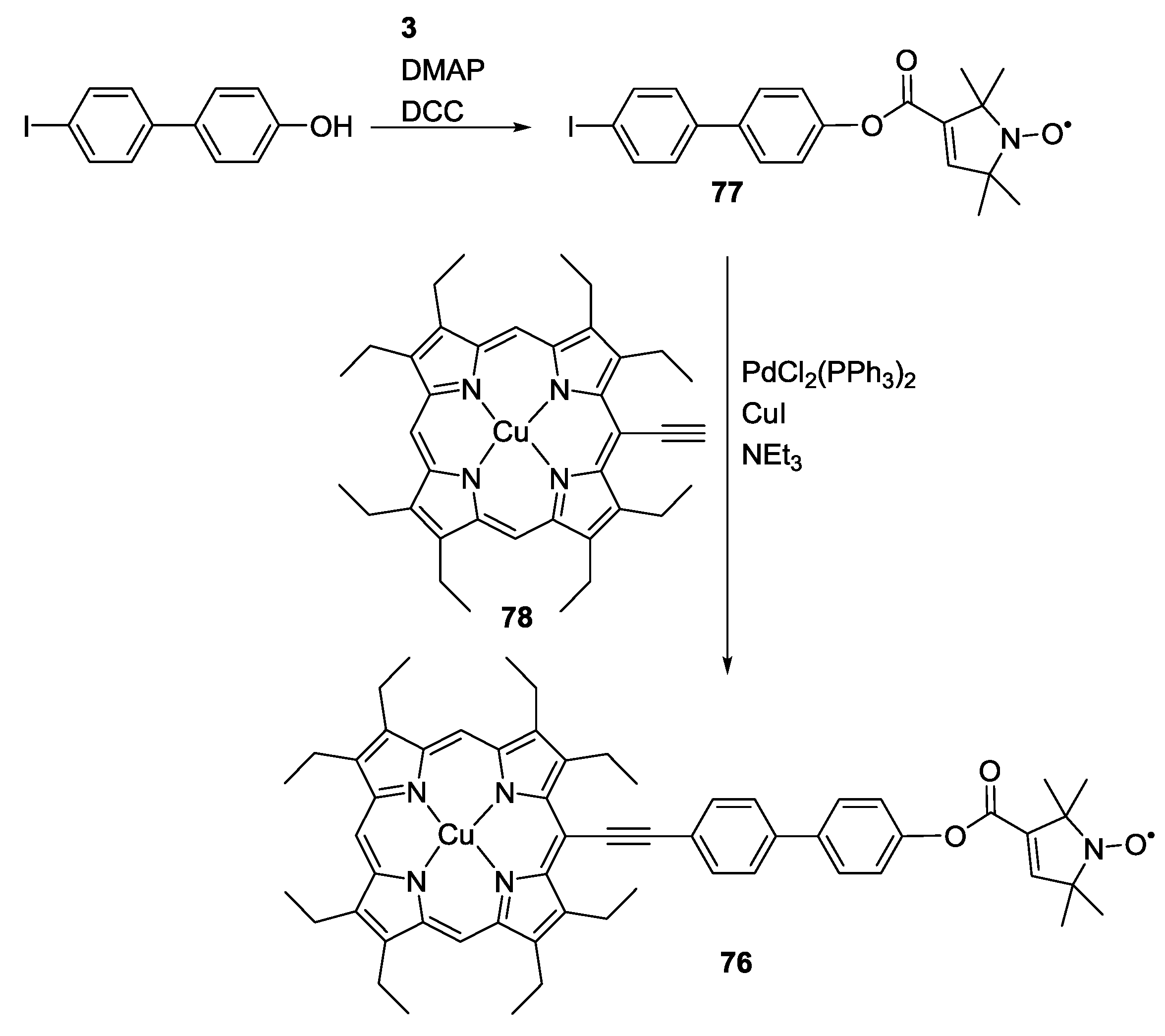
3.3. Metal Ions
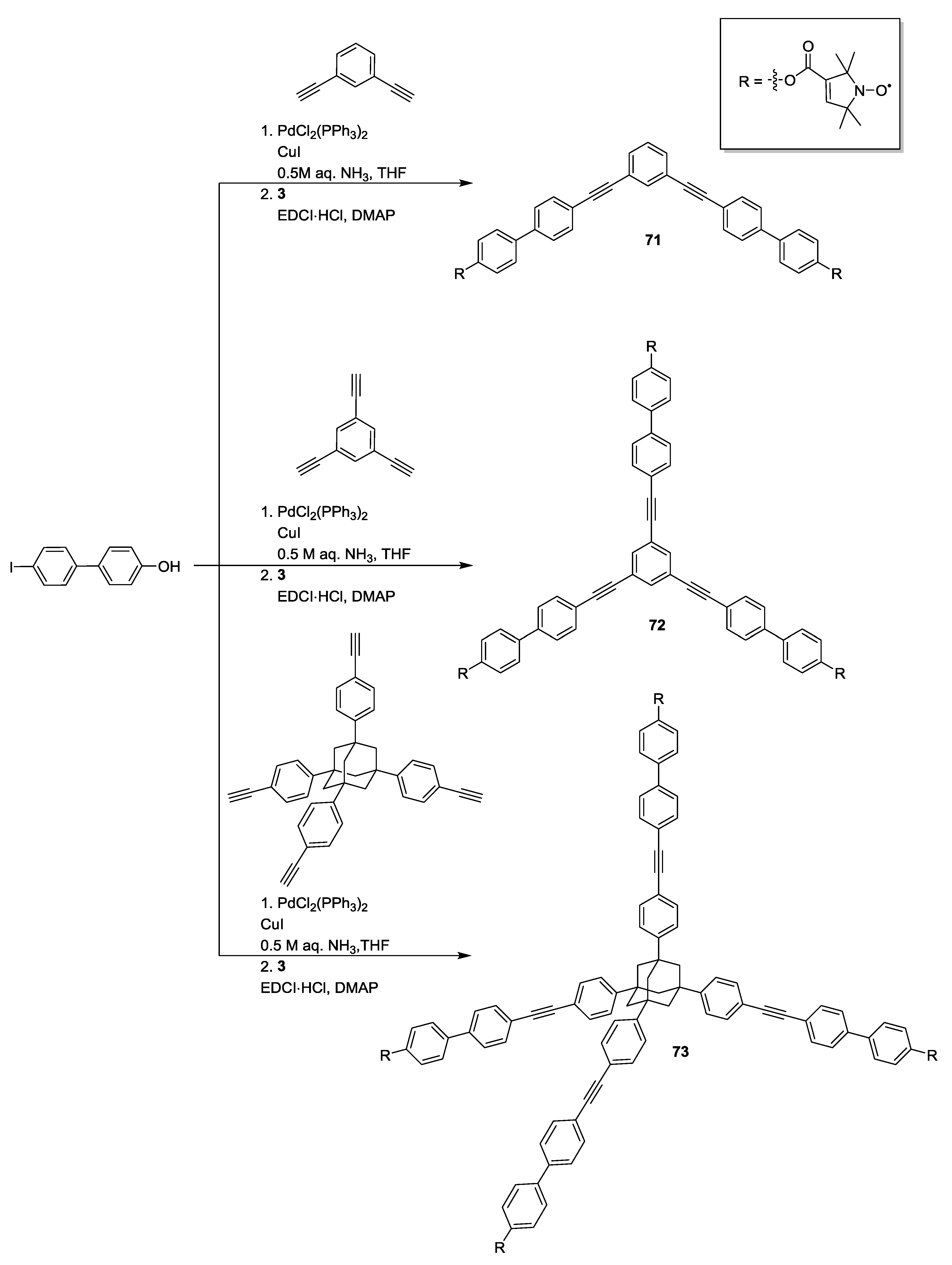
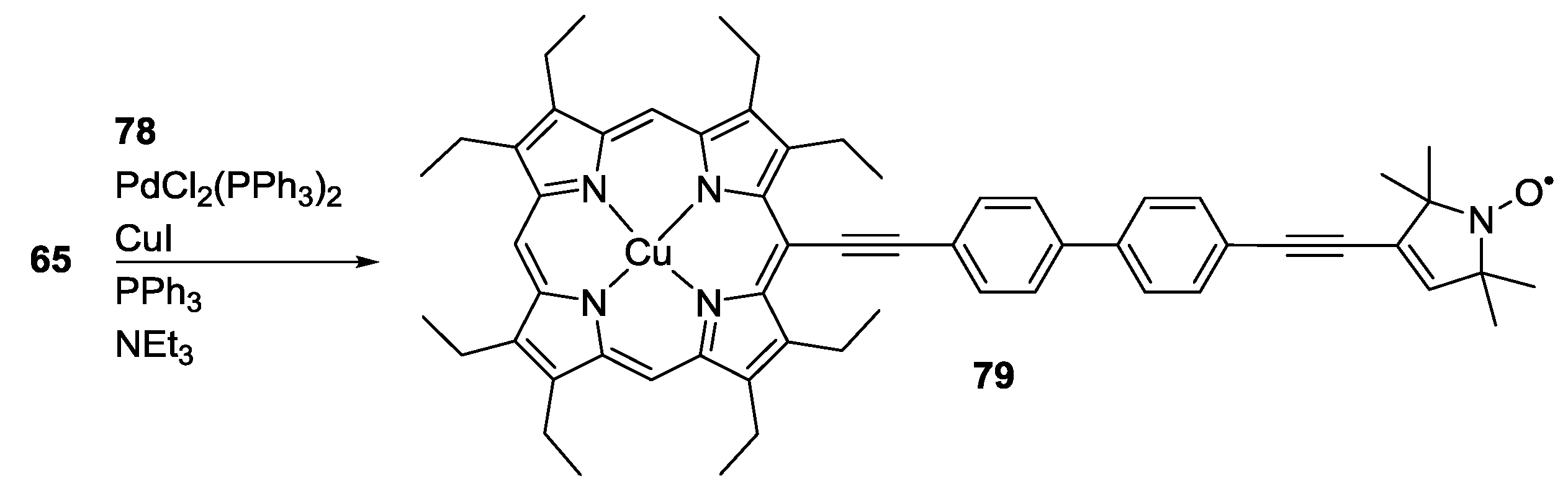
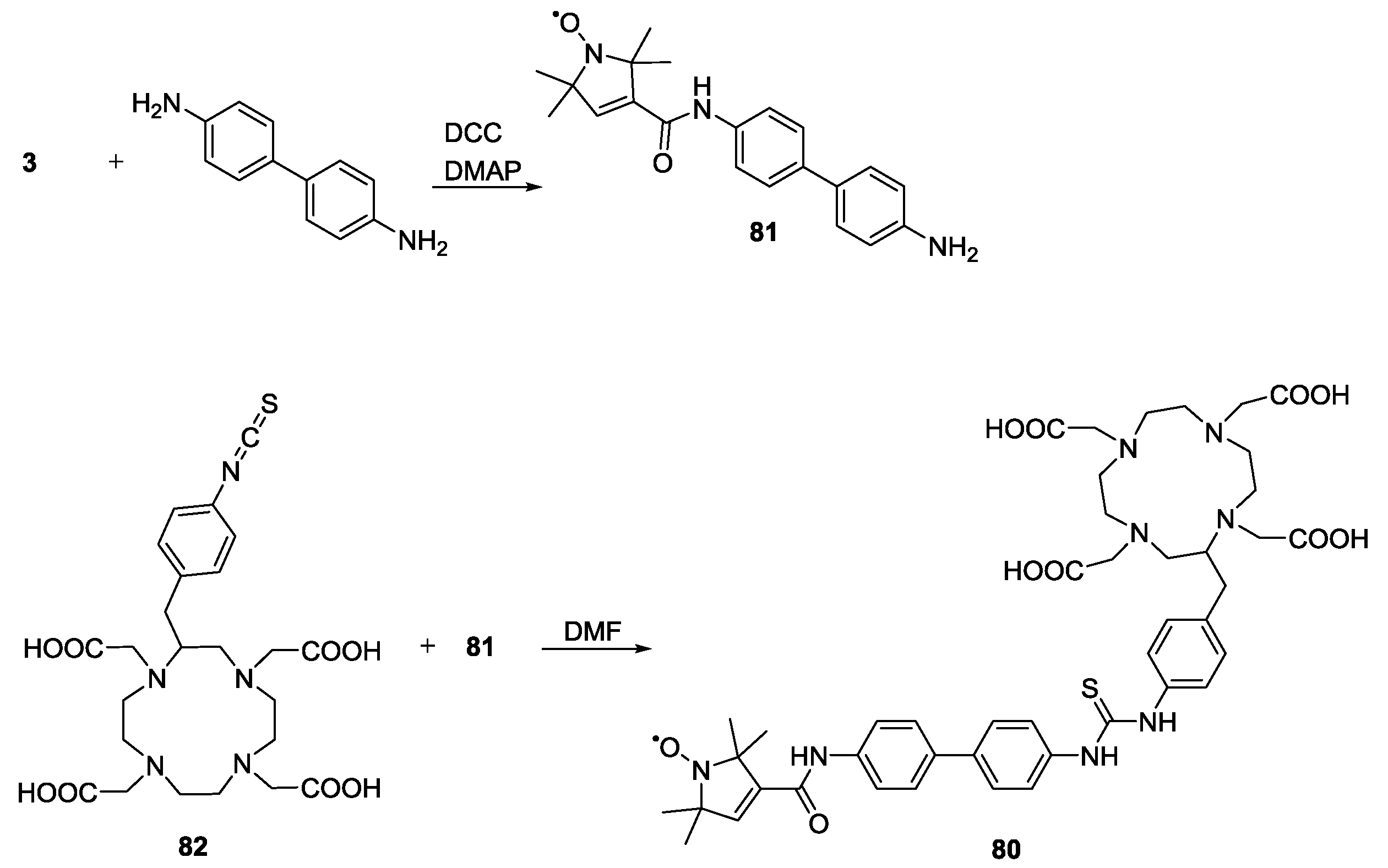

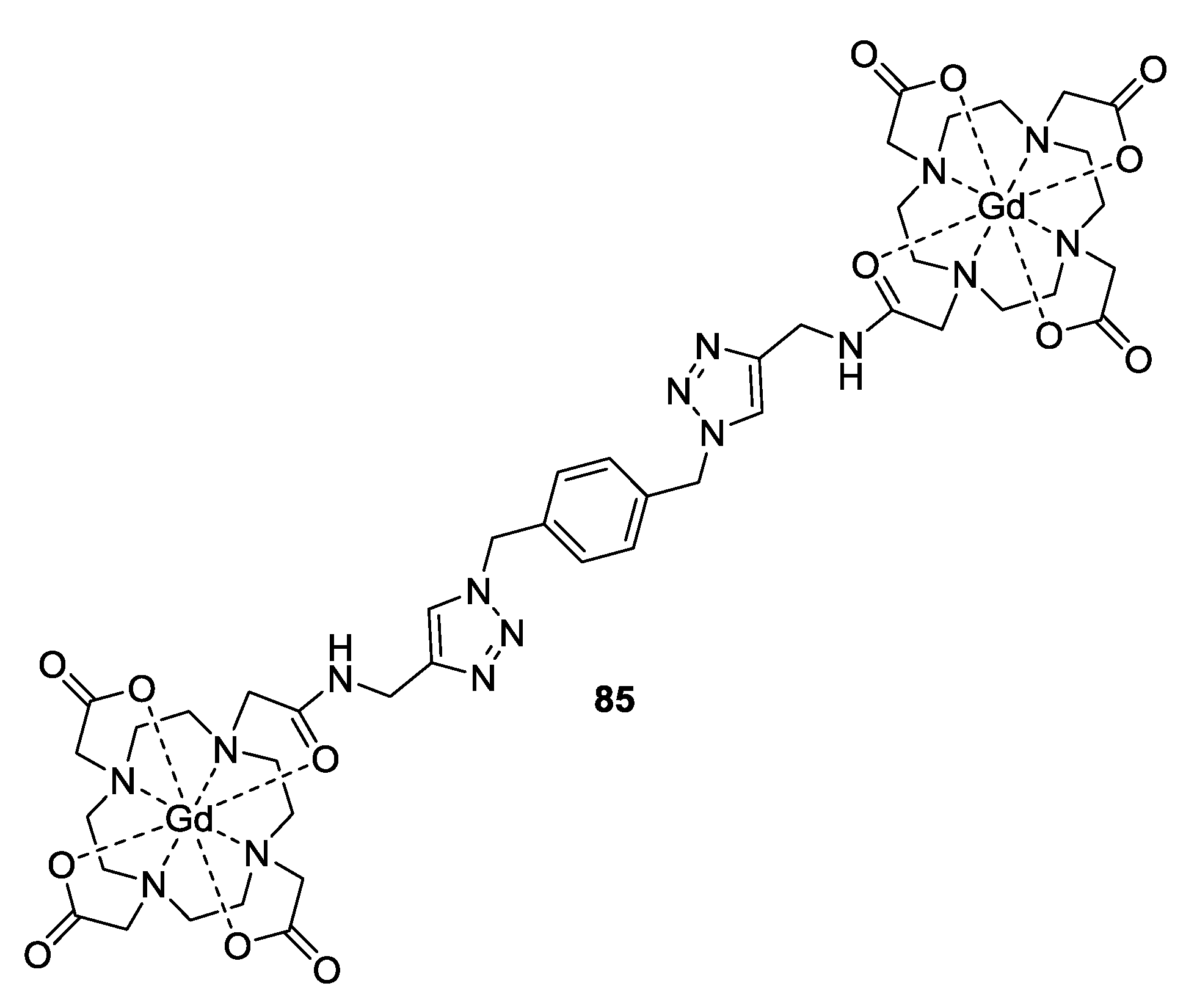
4. Applications
4.1. Polymer Degradation Studies via Distance Measurements

4.2. Biradicals as DNP NMR Polarising Agents
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schweiger, A. Pulsed electron spin resonance spectroscopy: Basic principles, techniques, and examples of applications. Angew. Chem. Int. Ed. Engl. 1991, 30, 265–292. [Google Scholar] [CrossRef]
- Schweiger, A.; Jeschke, G. Principles of Pulse Electron Paramagnetic Resonance; Oxford University Press: New York, NY, USA, 2001. [Google Scholar]
- Schiemann, O.; Prisner, T.F. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q. Rev. Biophys. 2007, 40, 1–53. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, G. DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 2012, 63, 419–446. [Google Scholar] [CrossRef] [PubMed]
- Elsässer, C.; Brecht, M.; Bittl, R. Pulsed electron−electron double resonance on multinuclear metal clusters: Assignment of spin projection factors based on the dipolar interaction. J. Am. Chem. Soc. 2002, 124, 12606–12611. [Google Scholar] [CrossRef] [PubMed]
- Mino, H.; Kawamori, A.; Ono, T.A. Pulsed EPR studies of doublet signal and singlet-like signal in oriented Ca2+-depleted PS II membranes: location of the doublet signal center in PS II. Biochemistry 2000, 39, 11034–11040. [Google Scholar] [CrossRef] [PubMed]
- Bennati, M.; Weber, A.; Antonic, J.; Perlstein, D.L.; Robblee, J.; Stubbe, J. Pulsed ELDOR spectroscopy measures the distance between the two tyrosyl radicals in the R2 subunit of the E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2003, 125, 14988–14989. [Google Scholar] [CrossRef] [PubMed]
- Cafiso, D.S.; Hubbell, W.L. EPR determination of membrane potentials. Annu. Rev. Biophys. Bioeng. 1981, 10, 217–244. [Google Scholar] [CrossRef] [PubMed]
- Hubbell, W.L.; Altenbach, C. Investigation of structure and dynamics in membrane proteins using site-directed spin labeling. Curr. Opin. Struct. Biol 1994, 4, 566–573. [Google Scholar] [CrossRef]
- Milov, A.D.; Ponomarev, A.B.; Tsvetkov, Y.D. Modulation beats of signal of double electron-electron resonance in spin echo for biradical systems. J. Struct. Chem. 1984, 25, 710–713. [Google Scholar] [CrossRef]
- Milov, A.D.; Maryasov, A.G.; Tsvetkov, Y.D. Pulsed electron double resonance (PELDOR) and its applications in free-radicals research. Appl. Magn. Reson. 1998, 15, 107–143. [Google Scholar] [CrossRef]
- Reginsson, G.W.; Shelke, S.A.; Rouillon, C.; White, M. F.; Sigurdsson, S. Th.; Schiemann, O. Protein-induced changes in DNA structure and dynamics observed with noncovalent site-directed spin labeling and PELDOR. Nucleic Acids Res. 2012, 41, e11–e21. [Google Scholar] [CrossRef] [PubMed]
- Milov, A.D.; Salikhov, K.M.; Shirov, M.D. Application of the double resonance method to electron spin echo in a study of the spatial distribution of paramagnetic centres in solids. Sov. Phys. Solid State 1981, 23, 565–569. [Google Scholar]
- Borbat, P.P.; Freed, J.H. Multiple-quantum ESR and distance measurements. Chem. Phys. Lett. 1999, 313, 145–154. [Google Scholar] [CrossRef]
- Freed, J.H. New technologies in electron spin resonance techniques. Annu. Rev. Phys. Chem. 2000, 51, 655–689. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Freed, J.H. Double quantum two-dimensional Fourier transform electron spin resonance: Distance measurements. Chem. Phys. Lett. 1996, 251, 102–110. [Google Scholar] [CrossRef]
- Rabenstein, M.D.; Shin, Y.K. Determination of the distance between two spin labels attached to a macromolecule. Proc. Natl. Acad. Sci. USA 1995, 92, 8239–8243. [Google Scholar] [CrossRef] [PubMed]
- Hustedt, E.; Beth, A. Structural Information from CW-EPR Spectra of Dipolar Coupled Nitroxide Spin Labels. In Distance Measurements in Biological Systems by EPR; Berliner, L., Eaton, G., Eaton, S., Eds.; Springer: New York, NY, USA, 2002; Volume 19, pp. 155–184. [Google Scholar]
- Jeschke, G.; Polyhach, Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys. Chem. Chem. Phys. 2007, 9, 1895–1910. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, Y.; Borbat, P.; Zweier, J.L.; Freed, J.H.; Hubbell, W.L. Pulsed ESR dipolar spectroscopy for distance measurements in immobilized spin labeled proteins in liquid solution. J. Am. Chem. Soc. 2012, 134, 9950–9952. [Google Scholar] [CrossRef] [PubMed]
- Larsen, R.G.; Singel, D.J. Double electron–electron resonance spin–echo modulation: Spectroscopic measurement of electron spin pair separations in orientationally disordered solids. J. Chem. Phys. 1993, 98, 5134–5146. [Google Scholar] [CrossRef]
- Marko, A.; Margraf, D.; Yu, H.; Mu, Y.; Stock, G.; Prisner, T. Molecular orientation studies by pulsed electron-electron double resonance experiments. J. Chem. Phys. 2009, 130, 064102. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, G.; Sajid, M.; Schulte, M.; Ramezanian, N.; Volkov, A.; Zimmermann, H.; Godt, A. Flexibility of shape-persistent molecular building blocks composed of p-phenylene and ethynylene units. J. Am. Chem. Soc. 2010, 132, 10107–10117. [Google Scholar] [CrossRef] [PubMed]
- Margraf, D.; Bode, B.E.; Marko, A.; Schiemann, O.; Prisner, T.F. Conformational flexibility of nitroxide biradicals determined by X-band PELDOR experiments. Mol. Phys. 2007, 105, 2153–2160. [Google Scholar] [CrossRef]
- Godt, A.; Schulte, M.; Zimmermann, H.; Jeschke, G. How flexible are poly(para-phenyleneethynylene)s? Angew. Chem. Int. Ed. 2006, 45, 7560–7564. [Google Scholar] [CrossRef]
- Jeschke, G.; Sajid, M.; Schulte, M.; Godt, A. Three-spin correlations in double electron-electron resonance. Phys. Chem. Chem. Phys. 2009, 11, 6580–6591. [Google Scholar] [CrossRef] [PubMed]
- Bode, B.E.; Margraf, D.; Plackmeyer, J.; Dürner, G.; Prisner, T.F.; Schiemann, O. Counting the monomers in nanometer-sized oligomers by pulsed electron−electron double resonance. J. Am. Chem. Soc. 2007, 129, 6736–6745. [Google Scholar] [CrossRef] [PubMed]
- Hilger, D.; Jung, H.; Padan, E.; Wegener, C.; Vogel, K.P.; Steinhoff, H.J.; Jeschke, G. Assessing oligomerization of membrane proteins by four-pulse DEER: pH-dependent dimerization of NhaA Na+/H+ antiporter of E. coli. Biophys. J. 2005, 89, 1328–1338. [Google Scholar] [CrossRef] [PubMed]
- Milov, A.D.; Ponomarev, A.B.; Tsvetkov, Y.D. Electron-electron double resonance in electron spin echo: Model biradical systems and the sensitized photolysis of decalin. Chem. Phys. Lett. 1984, 110, 67–72. [Google Scholar] [CrossRef]
- Margraf, D.; Cekan, P.; Prisner, T.F.; Sigurdsson, S.T.; Schiemann, O. Ferro- and antiferromagnetic exchange coupling constants in PELDOR spectra. Phys. Chem. Chem. Phys. 2009, 11, 6708–6714. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Schiemann, O.; Bode, B.; Prisner, T.F. PELDOR at S- and X-band frequencies and the separation of exchange coupling from dipolar coupling. J. Magn. Reson. 2002, 157, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, Y.D. Nitroxyls and PELDOR: Nitroxyl radicals in pulsed electron-electron double resonance spectroscopy. J. Struct. Chem. 2013, 54, 42–72. [Google Scholar] [CrossRef]
- Shelke, S.; Sigurdsson, S. Site-directed nitroxide spin labeling of biopolymers. In Structural Information from Spin-Labels and Intrinsic Paramagnetic Centres in the Biosciences; Timmel, C.R., Harmer, J.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 152, pp. 121–162. [Google Scholar]
- Reginsson, G.W.; Schiemann, O. Pulsed electron–electron double resonance: Beyond nanometre distance measurements on biomacromolecules. Biochem. J. 2011, 434, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, D. Metal-Based Spin Labeling for Distance Determination. In Structural Information from Spin-Labels and Intrinsic Paramagnetic Centres in the Biosciences; Timmel, C.R., Harmer, J.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 152, pp. 163–204. [Google Scholar]
- Altenbach, C.; Flitsch, S.L.; Khorana, H.G.; Hubbell, W.L. Structural studies on transmembrane proteins. 2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry 1989, 28, 7806–7812. [Google Scholar] [CrossRef] [PubMed]
- Valera, S.; Taylor, J.E.; Daniels, D.S.B.; Dawson, D.M.; Athukorala Arachchige, K.S.; Ashbrook, S.E.; Slawin, A.M.Z.; Bode, B.E. A modular approach for the synthesis of nanometer-sized polynitroxide multi-spin systems. J. Org. Chem. 2014, 79, 8313–8323. [Google Scholar] [CrossRef] [PubMed]
- Godt, A.; Franzen, C.; Veit, S.; Enkelmann, V.; Pannier, M.; Jeschke, G. EPR probes with well-defined, long distances between two or three unpaired electrons. J. Org. Chem. 2000, 65, 7575–7582. [Google Scholar] [CrossRef] [PubMed]
- Paletta, J.T.; Pink, M.; Foley, B.; Rajca, S.; Rajca, A. Synthesis and reduction kinetics of sterically shielded pyrrolidine nitroxides. Org. Lett. 2012, 14, 5322–5325. [Google Scholar] [CrossRef] [PubMed]
- Reginsson, G.W.; Kunjir, N.C.; Sigurdsson, S.T.; Schiemann, O. Trityl radicals: Spin labels for nanometer-distance measurements. Chem. Eur. J. 2012, 18, 13580–13584. [Google Scholar] [CrossRef] [PubMed]
- Martorana, A.; Bellapadrona, G.; Feintuch, A.; Di Gregorio, E.; Aime, S.; Goldfarb, D. Probing protein conformation in cells by EPR distance measurements using Gd3+ spin labeling. J. Am. Chem. Soc. 2014, 136, 13458–13465. [Google Scholar] [CrossRef] [PubMed]
- Pannier, M.; Veit, S.; Godt, A.; Jeschke, G.; Spiess, H.W. Dead-time free measurement of dipole–dipole interactions between electron spins. J. Magn. Reson. 2000, 142, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.E.; Pannier, M.; Diederich, F.; Gramlich, V.; Hubrich, M.; Spiess, H.W. Determination of end-to-end distances in a series of TEMPO diradicals of up to 2.8 nm length with a new four-pulse double electron electron resonance experiment. Angew. Chem. Int. Ed. 1998, 37, 2833–2837. [Google Scholar] [CrossRef]
- Jeschke, G.; Pannier, M.; Godt, A.; Spiess, H.W. Dipolar spectroscopy and spin alignment in electron paramagnetic resonance. Chem. Phys. Lett. 2000, 331, 243–252. [Google Scholar] [CrossRef]
- Borbat, P.P.; Freed, J.H. Measuring distances by pulsed dipolar ESR spectroscopy: Spin-labeled histidine kinases. Methods Enzymol. 2007, 423, 52–116. [Google Scholar] [PubMed]
- Schwab, P.F.H.; Levin, M.D.; Michl, J. Molecular rods. 1. Simple axial rods. Chem. Rev. 1999, 99, 1863–1933. [Google Scholar] [CrossRef] [PubMed]
- Pfannebecker, V.; Klos, H.; Hubrich, M.; Volkmer, T.; Heuer, A.; Wiesner, U.; Spiess, H.W. Determination of end-to-end distances in oligomers by pulsed EPR. J. Phys. Chem. 1996, 100, 13428–13432. [Google Scholar] [CrossRef]
- Potapov, A.; Song, Y.; Meade, T.J.; Goldfarb, D.; Astashkin, A.V.; Raitsimring, A. Distance measurements in model bis-Gd(III) complexes with flexible “bridge”. Emulation of biological molecules having flexible structure with Gd(III) labels attached. J. Magn. Reson. 2010, 205, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Raitsimring, A.M.; Gunanathan, C.; Potapov, A.; Efremenko, I.; Martin, J.M.L.; Milstein, D.; Goldfarb, D. Gd3+ complexes as potential spin labels for high field pulsed EPR distance measurements. J. Am. Chem. Soc. 2007, 129, 14138–14139. [Google Scholar] [CrossRef] [PubMed]
- Bode, B.E.; Plackmeyer, J.; Prisner, T.F.; Schiemann, O. PELDOR measurements on a nitroxide-labeled Cu(II) porphyrin: Orientation selection, spin-density distribution, and conformational flexibility. J. Phys. Chem. A 2008, 112, 5064–5073. [Google Scholar] [CrossRef] [PubMed]
- Narr, E.; Godt, A.; Jeschke, G. Selective measurements of a nitroxide–nitroxide separation of 5 nm and a nitroxide–copper separation of 2.5 nm in a terpyridine-based copper(II) complex by pulse EPR spectroscopy. Angew. Chem. Int. Ed. 2002, 41, 3907–3910. [Google Scholar] [CrossRef]
- Ezhevskaya, M.; Bordignon, E.; Polyhach, Y.; Moens, L.; Dewilde, S.; Jeschke, G.; Van Doorslaer, S. Distance determination between low-spin ferric haem and nitroxide spin label using DEER: The neuroglobin case. Mol. Phys. 2013, 111, 2855–2864. [Google Scholar] [CrossRef]
- Pornsuwan, S.; Bird, G.; Schafmeister, C.E.; Saxena, S. Flexibility and lengths of bis-peptide nanostructures by electron spin resonance. J. Am. Chem. Soc. 2006, 128, 3876–3877. [Google Scholar] [CrossRef] [PubMed]
- Schiemann, O.; Piton, N.; Mu, Y.; Stock, G.; Engels, J.W.; Prisner, T.F. A PELDOR-based nanometer distance ruler for oligonucleotides. J. Am. Chem. Soc. 2004, 126, 5722–5729. [Google Scholar] [CrossRef] [PubMed]
- Borbat, P.P.; Davis, J.H.; Butcher, S.E.; Freed, J.H. Measurement of large distances in biomolecules using double-quantum filtered refocused electron spin−echoes. J. Am. Chem. Soc. 2004, 126, 7746–7747. [Google Scholar] [CrossRef] [PubMed]
- Kunjir, N.C.; Reginsson, G.W.; Schiemann, O.; Sigurdsson, S.T. Measurements of short distances between trityl spin labels with CW EPR, DQC and PELDOR. Phys. Chem. Chem. Phys. 2013, 15, 19673–19685. [Google Scholar] [CrossRef] [PubMed]
- Grigor'ev, I.A.; Dikanov, S.A.; Shchukin, G.I.; Volodarskii, L.B.; Tsvetkov, Y.D. Influence of the N-oxide group in biradicals of the imidazoline series on the intramolecular spin exchange over systems of conjugated bonds. J. Struct. Chem. 1983, 23, 870–875. [Google Scholar] [CrossRef]
- Kulik, L.V.; Grishin, Y.A.; Dzuba, S.A.; Grigoryev, I.A.; Klyatskaya, S.V.; Vasilevsky, S.F.; Tsvetkov, Y.D. Electron dipole–dipole ESEEM in field-step ELDOR of nitroxide biradicals. J. Magn. Reson. 2002, 157, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Kulik, L.V.; Dzuba, S.A.; Grigoryev, I.A.; Tsvetkov, Y.D. Electron dipole–dipole interaction in ESEEM of nitroxide biradicals. Chem. Phys. Lett. 2001, 343, 315–324. [Google Scholar] [CrossRef]
- Kulik, L.V.; Paschenko, S.V.; Dzuba, S.A. 130 GHz ESEEM induced by electron–electron interaction in biradical. J. Magn. Reson. 2002, 159, 237–241. [Google Scholar] [CrossRef]
- Kulik, L.V.; Dzyuba, S.A. Electron spin echo study of molecular structure and dynamics: new approaches based on spontaneous fluctuations of magnetic interactions. J. Struct. Chem. 2004, 45, 298–314. [Google Scholar] [CrossRef]
- Rosantev, E.G. Free Nitroxide Radicals, 1st ed.; Press, P., Ed.; Plenum Press: New York, NY, USA, 1970; p. 213. [Google Scholar]
- Jeschke, G.; Panek, G.; Godt, A.; Bender, A.; Paulsen, H. Data analysis procedures for pulse ELDOR measurements of broad distance distributions. Appl. Magn. Reson. 2004, 26, 223–244. [Google Scholar] [CrossRef]
- Neises, B.; Steglich, W. Simple method for the esterification of carboxylic acids. Angew. Chem. Int. Ed. 1978, 17, 522–524. [Google Scholar] [CrossRef]
- Jeschke, G.; Zimmermann, H.; Godt, A. Isotope selection in distance measurements between nitroxides. J. Magn. Reson. 2006, 180, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, G.; Chechik, V.; Ionita, P.; Godt, A.; Zimmermann, H.; Banham, J.; Timmel, C.R.; Hilger, D.; Jung, H. DeerAnalysis2006—A comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006, 30, 473–498. [Google Scholar] [CrossRef]
- Jeschke, G.; Koch, A.; Jonas, U.; Godt, A. Direct conversion of EPR dipolar time evolution data to distance distributions. J. Magn. Reson. 2002, 155, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, G.; Bender, A.; Paulsen, H.; Zimmermann, H.; Godt, A. Sensitivity enhancement in pulse EPR distance measurements. J. Magn. Reson. 2004, 169, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.; Jeschke, G.; Wiebcke, M.; Godt, A. Conformationally unambiguous spin labeling for distance measurements. Chem. Eur. J. 2009, 15, 12960–12962. [Google Scholar] [CrossRef] [PubMed]
- Marko, A.; Denysenkov, V.; Margraf, D.; Cekan, P.; Schiemann, O.; Sigurdsson, S.T.; Prisner, T.F. Conformational flexibility of DNA. J. Am. Chem. Soc. 2011, 133, 13375–13379. [Google Scholar] [CrossRef] [PubMed]
- Marko, A.; Prisner, T.F. An algorithm to analyze PELDOR data of rigid spin label pairs. Phys. Chem. Chem. Phys. 2013, 15, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Marko, A.; Denysenkov, V.; Prisner, T.F. Out-of-phase PELDOR. Mol. Phys. 2013, 111, 2834–2844. [Google Scholar] [CrossRef]
- Spindler, P.E.; Glaser, S.J.; Skinner, T.E.; Prisner, T.F. Broadband inversion PELDOR spectroscopy with partially adiabatic shaped pulses. Angew. Chem. Int. Ed. 2013, 52, 3425–3429. [Google Scholar] [CrossRef]
- Schiemann, O.; Cekan, P.; Margraf, D.; Prisner, T.F.; Sigurdsson, S.T. Relative orientation of rigid nitroxides by PELDOR: Beyond distance measurements in nucleic acids. Angew. Chem. Int. Ed. 2009, 48, 3292–3295. [Google Scholar] [CrossRef]
- Reginsson, G.W.; Hunter, R.I.; Cruickshank, P.A.S.; Bolton, D.R.; Sigurdsson, S.T.; Smith, G.M.; Schiemann, O. W-band PELDOR with 1kW microwave power: molecular geometry, flexibility and exchange coupling. J. Magn. Reson. 2012, 216, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Bonora, M.; Becker, J.; Saxena, S. Suppression of electron spin-echo envelope modulation peaks in double quantum coherence electron spin resonance. J. Magn. Reson. 2004, 170, 278–283. [Google Scholar] [CrossRef] [PubMed]
- von Hagens, T.; Polyhach, Y.; Sajid, M.; Godt, A.; Jeschke, G. Suppression of ghost distances in multiple-spin double electron-electron resonance. Phys. Chem. Chem. Phys. 2013, 15, 5854–5866. [Google Scholar] [CrossRef] [PubMed]
- Giannoulis, A.; Ward, R.; Branigan, E.; Naismith, J.H.; Bode, B.E. PELDOR in rotationally symmetric homo-oligomers. Mol. Phys. 2013, 111, 2845–2854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polyhach, Y.; Godt, A.; Bauer, C.; Jeschke, G. Spin pair geometry revealed by high-field DEER in the presence of conformational distributions. J. Magn. Reson. 2007, 185, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Polyhach, Y.; Bordignon, E.; Tschaggelar, R.; Gandra, S.; Godt, A.; Jeschke, G. High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Phys. Chem. Chem. Phys. 2012, 14, 10762–10773. [Google Scholar] [CrossRef] [PubMed]
- Kay, C.W.M.; El Mkami, H.; Cammack, R.; Evans, R.W. Pulsed ELDOR determination of the intramolecular distance between the metal binding sites in dicupric human serum transferrin and lactoferrin. J. Am. Chem. Soc. 2007, 129, 4868–4869. [Google Scholar] [CrossRef] [PubMed]
- Van Amsterdam, I.M.C.; Ubbink, M.; Canters, G.W.; Huber, M. Measurement of a Cu-Cu distance of 26 A by a pulsed EPR method. Angew. Chem. Int. Ed. 2003, 42, 62–64. [Google Scholar] [CrossRef]
- Van Wonderen, J.H.; Kostrz, D.N.; Dennison, C.; MacMillan, F. Refined distances between paramagnetic centers of a multi-copper nitrite reductase determined by pulsed EPR (iDEER) spectroscopy. Angew. Chem. Int. Ed. 2013, 52, 1990–1993. [Google Scholar] [CrossRef]
- Denysenkov, V.P.; Prisner, T.F.; Stubbe, J.; Bennati, M. High-field pulsed electron–electron double resonance spectroscopy to determine the orientation of the tyrosyl radicals in ribonucleotide reductase. Proc. Natl. Acad. Sci. USA 2006, 103, 13386–13390. [Google Scholar] [CrossRef] [PubMed]
- Eaton, S.; Eaton, G. Relaxation Times of Organic Radicals and Transition Metal Ions. In Distance Measurements in Biological Systems by EPR; Berliner, L., Eaton, G., Eaton, S., Eds.; Springer: New York, NY, USA, 2002; Volume 19, pp. 29–154. [Google Scholar]
- Bode, B.E.; Plackmeyer, J.; Bolte, M.; Prisner, T.F.; Schiemann, O. PELDOR on an exchange coupled nitroxide copper(II) spin pair. J. Organomet. Chem. 2009, 694, 1172–1179. [Google Scholar] [CrossRef]
- Jäger, H.; Koch, A.; Maus, V.; Spiess, H.W.; Jeschke, G. Relaxation-based distance measurements between a nitroxide and a lanthanide spin label. J. Magn. Reson. 2008, 194, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Lueders, P.; Jeschke, G.; Yulikov, M. Double electron−electron resonance measured between Gd3+ ions and nitroxide radicals. J. Phys. Chem. Lett. 2011, 2, 604–609. [Google Scholar] [CrossRef]
- Jeschke, G.; Schlick, S. Spatial distribution of stabilizer-derived nitroxide radicals during thermal degradation of poly(acrylonitrile-butadiene-styrene) copolymers: a unified picture from pulsed ELDOR and ESR imaging. Phys. Chem. Chem. Phys. 2006, 8, 4095–4103. [Google Scholar] [CrossRef] [PubMed]
- Hall, D.A.; Maus, D.C.; Gerfen, G.J.; Inati, S.J.; Becerra, L.R.; Dahlquist, F.W.; Griffin, R.G. Polarization-enhanced NMR spectroscopy of biomolecules in frozen solution. Science 1997, 276, 930–932. [Google Scholar] [CrossRef] [PubMed]
- Sauvée, C.; Rosay, M.; Casano, G.; Aussenac, F.; Weber, R.T.; Ouari, O.; Tordo, P. Highly efficient, water-soluble polarizing agents for dynamic nuclear polarization at high frequency. Angew. Chem. Int. Ed. 2013, 52, 10858–10861. [Google Scholar] [CrossRef]
- Matsuki, Y.; Maly, T.; Ouari, O.; Karoui, H.; Le Moigne, F.; Rizzato, E.; Lyubenova, S.; Herzfeld, J.; Prisner, T.; Tordo, P.; et al. Dynamic nuclear polarization with a rigid biradical. Angew. Chem. Int. Ed. 2009, 48, 4996–5000. [Google Scholar] [CrossRef]
- Kiesewetter, M.K.; Corzilius, B.; Smith, A.A.; Griffin, R.G.; Swager, T.M. Dynamic nuclear polarization with a water-soluble rigid biradical. J. Am. Chem. Soc. 2012, 134, 4537–4540. [Google Scholar] [CrossRef] [PubMed]
- Zagdoun, A.; Casano, G.; Ouari, O.; Schwarzwälder, M.; Rossini, A.J.; Aussenac, F.; Yulikov, M.; Jeschke, G.; Copéret, C.; Lesage, A.; et al. Large molecular weight nitroxide biradicals providing efficient dynamic nuclear polarization at temperatures up to 200 K. J. Am. Chem. Soc. 2013, 135, 12790–12797. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valera, S.; Bode, B.E. Strategies for the Synthesis of Yardsticks and Abaci for Nanometre Distance Measurements by Pulsed EPR. Molecules 2014, 19, 20227-20256. https://doi.org/10.3390/molecules191220227
Valera S, Bode BE. Strategies for the Synthesis of Yardsticks and Abaci for Nanometre Distance Measurements by Pulsed EPR. Molecules. 2014; 19(12):20227-20256. https://doi.org/10.3390/molecules191220227
Chicago/Turabian StyleValera, Silvia, and Bela E. Bode. 2014. "Strategies for the Synthesis of Yardsticks and Abaci for Nanometre Distance Measurements by Pulsed EPR" Molecules 19, no. 12: 20227-20256. https://doi.org/10.3390/molecules191220227