Molecular Docking Characterization of a Four-Domain Segment of Human Fibronectin Encompassing the RGD Loop with Hydroxyapatite
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Potential HAP-Binding Sites on FN-III7–10 Surface and Molecular Docking of HAP to FN

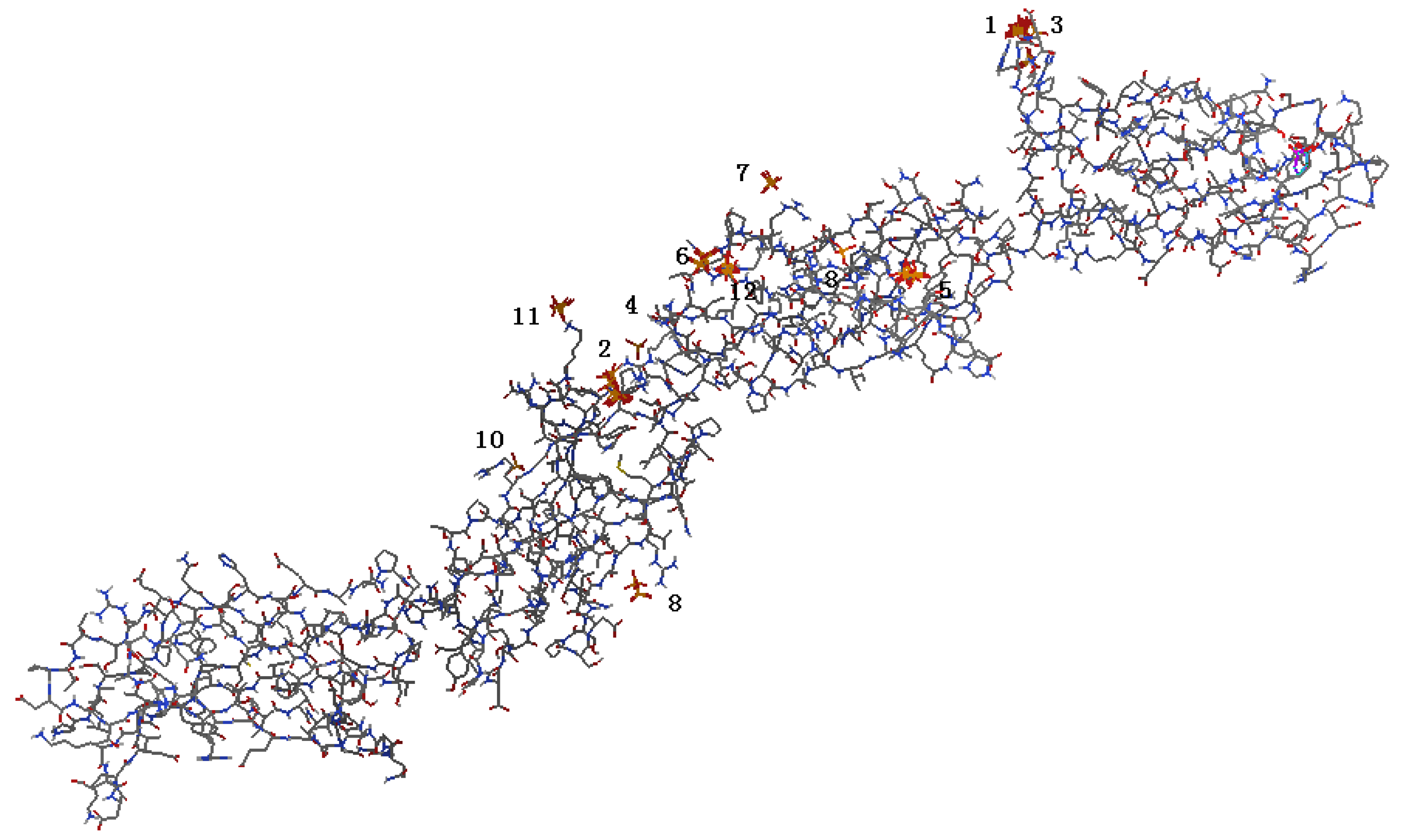


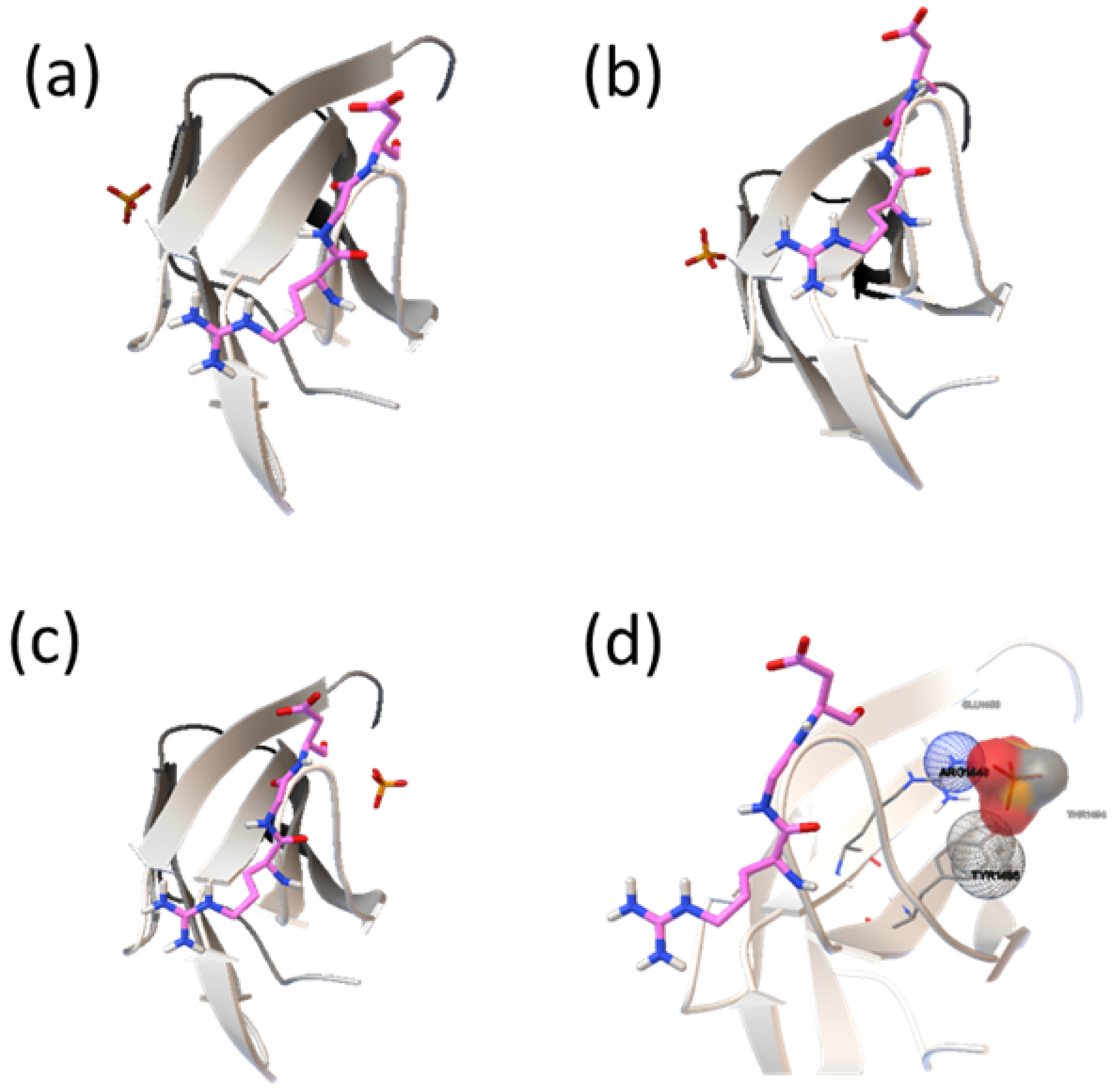
2.2. Analysis of HAP-Binding Site on RGD Loop
3. Experimental
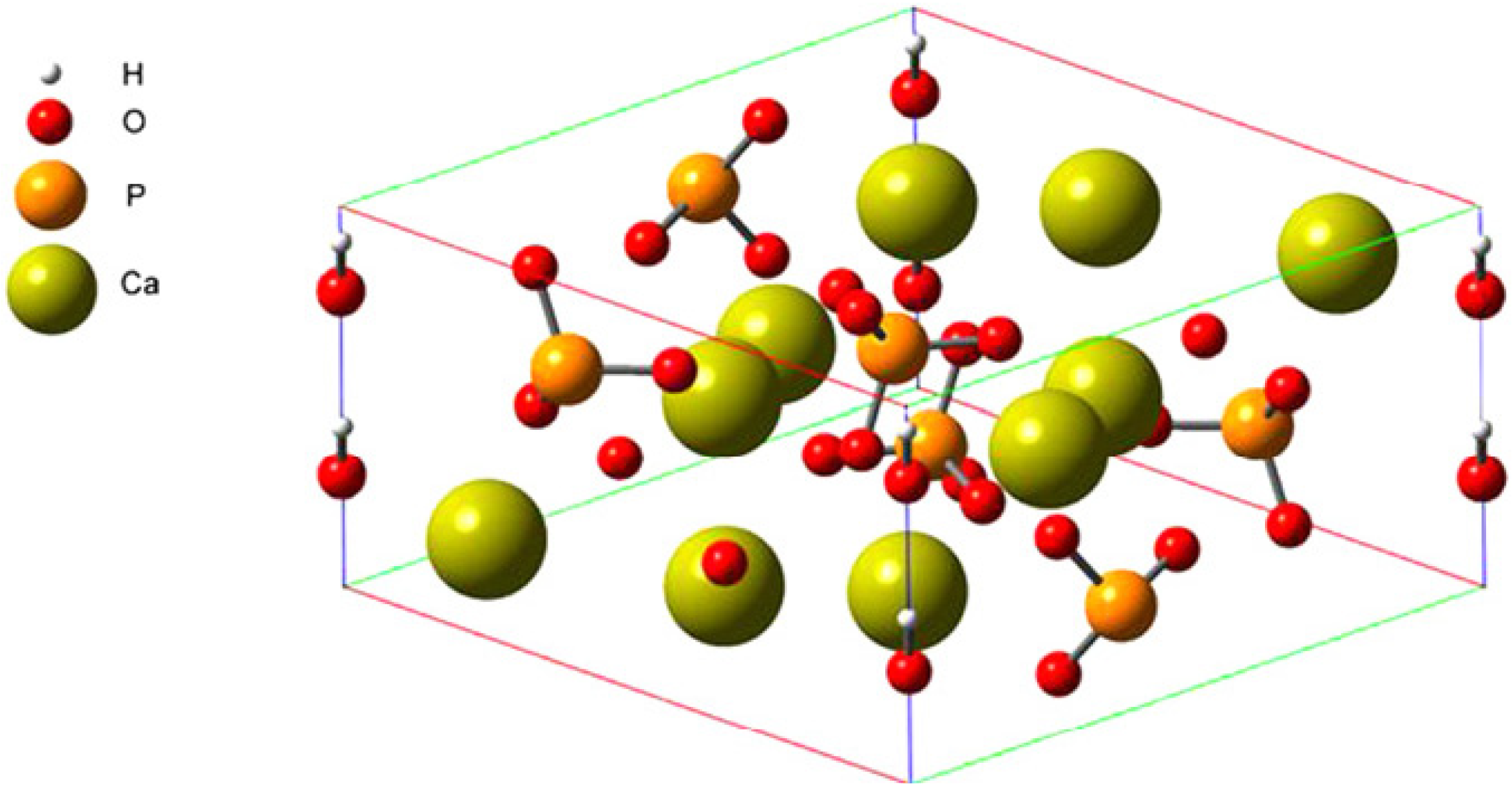
3.1. Setup of FN-III7–10 and HAP Structures

3.2. The FN-III7–10 Binding Site Prediction with MPK2
3.3. Performing Molecular Docking with AutoDock 4
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Romberger, D.J. Fibronectin. Int. J. Biochem. Cell Biol. 1997, 29, 939–943. [Google Scholar]
- Brumfeld, V.; Werber, M. Studies on fibronectin and its domains: II. Secondary structure and spatial configuration of fibronectin and of its domains. Arch. Biochem. Biophys. 1993, 302, 134–143. [Google Scholar] [CrossRef]
- Leahy, D.J.; Aukhil, I.; Erickson, H.P. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 1996, 84, 155–164. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- George, E.L.; Georges-Labouesse, E.N.; Patel-King, R.S.; Rayburn, H.; Hynes, R.O. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 1993, 119, 1079–1091. [Google Scholar]
- Pfaff, M.; McLane, M.A.; Beviglia, L.; Niewiarowski, S.; Timpl, R. Comparison of disintegrins with limited variation in the RGD loop in their binding to purified integrins αIIbβ3, αVβ3 and α5β1 and in cell adhesion inhibition. Cell Commun. Adh. 1994, 2, 491–501. [Google Scholar] [CrossRef]
- Dolatshahi-Pirouz, A.; Jensen, T.; Foss, M.; Chevallier, J.; Besenbacher, F. Enhanced surface activation of fibronectin upon adsorption on hydroxyapatite. Langmuir 2009, 25, 2971–2978. [Google Scholar] [CrossRef]
- Block, M.; Finger, I.; Fontenot, M.; Kent, J. Loaded hydroxylapatite-coated and grit-blasted titanium implants in dogs. Int. J. Oral Maxillofac. Implants 1989, 4, 219–225. [Google Scholar]
- Kasemo, B. Biological surface science. Surf. Sci. 2002, 500, 656–677. [Google Scholar] [CrossRef]
- Holmberg, K. Novel Surfactants: Preparation Applications and Biodegradability, Revised and Expanded, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- Amaral, I.F.; Lamghari, M.; Sousa, S.R.; Sampaio, P.; Barbosa, M.A. Rat bone marrow stromal cell osteogenic differentiation and fibronectin adsorption on chitosan membranes: The effect of the degree of acetylation. J. Biomed. Mater. Res. A 2005, 75, 387–397. [Google Scholar]
- Cairns, M.L.; Meenan, B.J.; Burke, G.A.; Boyd, A.R. Influence of surface topography on osteoblast response to fibronectin coated calcium phosphate thin films. Colloids Surf. B Biointerfaces 2010, 78, 283–290. [Google Scholar] [CrossRef]
- Docheva, D.; Padula, D.; Schieker, M.; Clausen-Schaumann, H. Effect of collagen I and fibronectin on the adhesion, elasticity and cytoskeletal organization of prostate cancer cells. Biochem. Biophys. Res. Commun. 2010, 402, 361–366. [Google Scholar] [CrossRef]
- Hindie, M.; Degat, M.C.; Gaudiere, F.; Gallet, O.; van Tassel, P.R.; Pauthe, E. Pre-osteoblasts on poly(l-lactic acid) and silicon oxide: Influence of fibronectin and albumin adsorption. Acta Biomater. 2011, 7, 387–394. [Google Scholar] [CrossRef]
- Geiger, B.; Bershadsky, A.; Pankov, R.; Yamada, K.M. Transmembrane crosstalk between the extracellular matrix and the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2001, 2, 793–805. [Google Scholar] [CrossRef]
- Wallwork, M.L.; Kirkham, J.; Zhang, J.; Smith, D.A.; Brookes, S.J.; Shore, R.C.; Wood, S.R.; Ryu, O.; Robinson, C. Binding of matrix proteins to developing enamel crystals: An atomic force microscopy study. Langmuir 2001, 17, 2508–2513. [Google Scholar]
- Kandori, K.; Murata, K.; Ishikawa, T. Microcalorimetric study of protein adsorption onto calcium hydroxyapatites. Langmuir 2007, 23, 2064–2070. [Google Scholar] [CrossRef]
- Gibson, J.M.; Popham, J.M.; Raghunathan, V.; Stayton, P.S.; Drobny, G.P. A solid-state NMR study of the dynamics and interactions of phenylalanine rings in a statherin fragment bound to hydroxyapatite crystals. J. Am. Chem. Soc. 2006, 128, 5364–5370. [Google Scholar] [CrossRef]
- Vitorino, R.; Lobo, M.J.C.; Duarte, J.; Ferrer-Correia, A.J.; Tomer, K.B.; Dubin, J.R.; Domingues, P.M.; Amado, F.M.L. In vitro hydroxyapatite adsorbed salivary proteins. Biochem. Biophys. Res. Commun. 2004, 320, 342–346. [Google Scholar] [CrossRef]
- Shen, J.W.; Wu, T.; Wang, Q.; Pan, H.H. Molecular simulation of protein adsorption and desorption on hydroxyapatite surfaces. Biomaterials 2008, 29, 513–532. [Google Scholar] [CrossRef]
- Huang, B. MetaPocket: A meta approach to improve protein ligand binding site prediction. OMICS 2009, 13, 325–330. [Google Scholar] [CrossRef]
- Lu, J.; Shi, M.; Shoichet, M.S. Click chemistry functionalized polymeric nanoparticles target corneal epithelial cells through RGD-cell surface receptors. Bioconjug. Chem. 2008, 20, 87–94. [Google Scholar]
- Aota, S.I.; Nomizu, M.; Yamada, K.M. The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell-adhesive function. J. Biol. Chem. 1994, 269, 24756–24761. [Google Scholar]
- Bowditch, R.D.; Hariharan, M.; Tominna, E.F.; Smith, J.W.; Yamada, K.M.; Getzoff, E.D.; Ginsberg, M.H. Identification of a novel integrin binding site in fibronectin. Differential utilization by beta 3 integrins. J. Biol. Chem. 1994, 269, 10856–10863. [Google Scholar]
- Riener, C.K.; Kienberger, F.; Hahn, C.D.; Buchinger, G.M.; Egwim, I.O.C.; Haselgrübler, T.; Ebner, A.; Romanin, C.; Klampfl, C.; Lackner, B.; et al. Heterobifunctional crosslinkers for tethering single ligand molecules to scanning probes. Anal. Chim. Acta 2003, 497, 101–114. [Google Scholar] [CrossRef]
- Vignoles, M.; Bonel, G.; Holcomb, D.; Young, R. Influence of preparation conditions on the composition of type B carbonated hydroxyapatite and on the localization of the carbonate ions. Calcif. Tissue Int. 1988, 43, 33–40. [Google Scholar] [CrossRef]
- Vignoles, M.; Bonel, G.; Young, R. Occurrence of nitrogenous species in precipitated B-type carbonated hydroxyapatites. Calcif. Tissue Int. 1987, 40, 64–70. [Google Scholar] [CrossRef]
- Sudarsanan, K.T.; Young, R. Significant precision in crystal structural details. Holly Springs hydroxyapatite. Acta Crystallogr. B 1969, 25, 1534–1543. [Google Scholar] [CrossRef]
- Feki, H.E.; Savariault, J.M.; Salah, A.B. Structure refinements by the Rietveld method of partially substituted hydroxyapatite: Ca9Na0.5(PO4)4.5(CO3)1.5(OH)2. J. Alloys Compd. 1999, 287, 114–120. [Google Scholar] [CrossRef]
- Menendez-Proupin, E.; Cervantes-Rodriguez, S.; Osorio-Pulgar, R.; Franco-Cisterna, M.; Camacho-Montes, H.; Fuentes, M.E. Computer simulation of elastic constants of hydroxyapatite and fluorapatite. J. Mech. Behav. Biomed. Mater. 2011, 4, 1011–1020. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Y.; Lin, B.; Schroeder, M.; Huang, B. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics 2011, 27, 2083–2088. [Google Scholar] [CrossRef]
- MPK2 Home Page. Available online: http://projects.biotec.tu-dresden.de/metapocket/ (accessed on 16 December 2003).
- Sousa, S.F.; Fernandes, P.A.; Ramos, M.J. Protein-ligand docking: Current status and future challenges. Proteins 2006, 65, 15–26. [Google Scholar] [CrossRef]
- Jiang, X.; Kumar, K.; Hu, X.; Wallqvist, A.; Reifman, J. DOVIS 2.0: An efficient and easy to use parallel virtual screening tool based on AutoDock 4.0. Chem. Cent. J. 2008, 2, 18. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2013 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Guo, T.; Kang, W.; Xiao, D.; Duan, R.; Zhi, W.; Weng, J. Molecular Docking Characterization of a Four-Domain Segment of Human Fibronectin Encompassing the RGD Loop with Hydroxyapatite. Molecules 2014, 19, 149-158. https://doi.org/10.3390/molecules19010149
Guo T, Kang W, Xiao D, Duan R, Zhi W, Weng J. Molecular Docking Characterization of a Four-Domain Segment of Human Fibronectin Encompassing the RGD Loop with Hydroxyapatite. Molecules. 2014; 19(1):149-158. https://doi.org/10.3390/molecules19010149
Chicago/Turabian StyleGuo, Tailin, Wenyuan Kang, Dongqin Xiao, Rongquan Duan, Wei Zhi, and Jie Weng. 2014. "Molecular Docking Characterization of a Four-Domain Segment of Human Fibronectin Encompassing the RGD Loop with Hydroxyapatite" Molecules 19, no. 1: 149-158. https://doi.org/10.3390/molecules19010149
APA StyleGuo, T., Kang, W., Xiao, D., Duan, R., Zhi, W., & Weng, J. (2014). Molecular Docking Characterization of a Four-Domain Segment of Human Fibronectin Encompassing the RGD Loop with Hydroxyapatite. Molecules, 19(1), 149-158. https://doi.org/10.3390/molecules19010149



